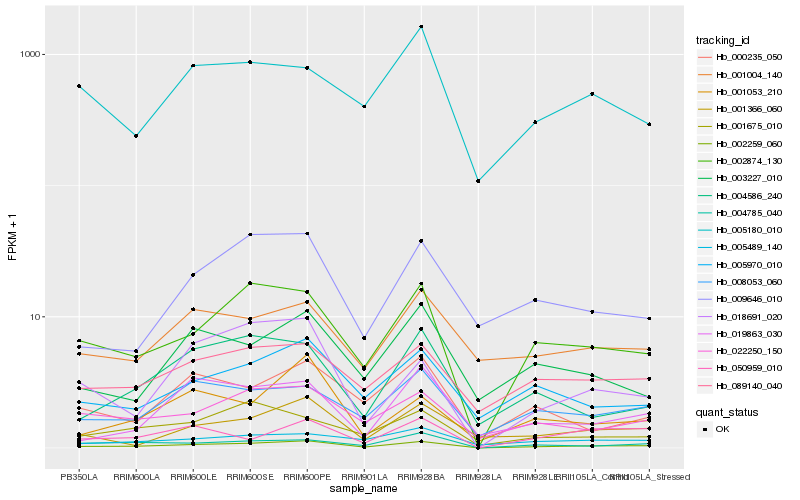
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002259_060 |
0.0 |
- |
- |
PREDICTED: kinesin-like calmodulin-binding protein isoform X4 [Jatropha curcas] |
| 2 |
Hb_002874_130 |
0.1209561351 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 9 [Jatropha curcas] |
| 3 |
Hb_022250_150 |
0.1640434488 |
- |
- |
PREDICTED: uncharacterized protein LOC105648687 [Jatropha curcas] |
| 4 |
Hb_008053_060 |
0.1648351454 |
- |
- |
PREDICTED: protein PAIR1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_004785_040 |
0.1686642852 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 3-like [Jatropha curcas] |
| 6 |
Hb_005970_010 |
0.1713266594 |
- |
- |
PREDICTED: probable isoprenylcysteine alpha-carbonyl methylesterase ICMEL1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_089140_040 |
0.1714158375 |
- |
- |
PREDICTED: uncharacterized protein LOC105637966 [Jatropha curcas] |
| 8 |
Hb_018691_020 |
0.1829089389 |
- |
- |
- |
| 9 |
Hb_001675_010 |
0.1834922118 |
- |
- |
hypothetical protein TRIUR3_09961 [Triticum urartu] |
| 10 |
Hb_004586_240 |
0.1858212458 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein At1g28390 [Jatropha curcas] |
| 11 |
Hb_005489_140 |
0.1872344209 |
- |
- |
PREDICTED: maspardin [Jatropha curcas] |
| 12 |
Hb_000235_050 |
0.1874567437 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001004_140 |
0.1874881242 |
- |
- |
hypothetical protein JCGZ_26893 [Jatropha curcas] |
| 14 |
Hb_050959_010 |
0.1879303014 |
- |
- |
- |
| 15 |
Hb_003227_010 |
0.188789307 |
- |
- |
PREDICTED: dynamin-related protein 1E-like [Jatropha curcas] |
| 16 |
Hb_001366_060 |
0.1888244793 |
- |
- |
- |
| 17 |
Hb_009646_010 |
0.1892598732 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase-like [Jatropha curcas] |
| 18 |
Hb_001053_210 |
0.190604402 |
- |
- |
- |
| 19 |
Hb_019863_030 |
0.1922678202 |
- |
- |
hypothetical protein JCGZ_00393 [Jatropha curcas] |
| 20 |
Hb_005180_010 |
0.1923311455 |
- |
- |
cyclophilin [Hevea brasiliensis] |