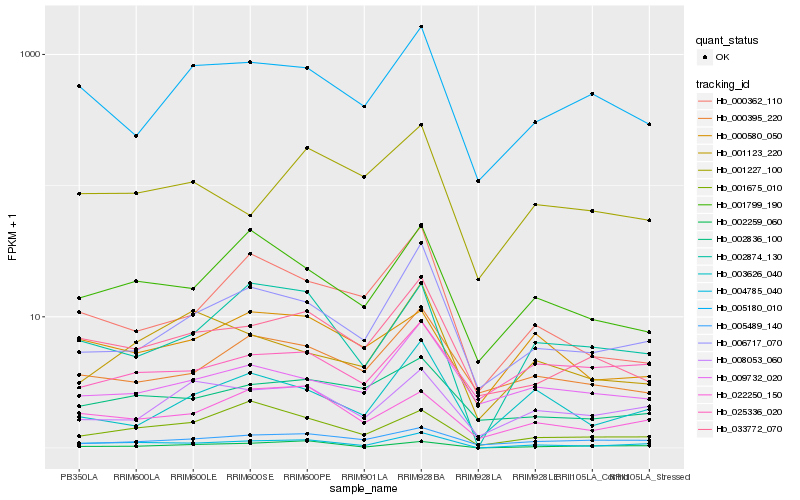
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004785_040 |
0.0 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 3-like [Jatropha curcas] |
| 2 |
Hb_002874_130 |
0.15957265 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 9 [Jatropha curcas] |
| 3 |
Hb_006717_070 |
0.1632577373 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor CPC-like [Jatropha curcas] |
| 4 |
Hb_033772_070 |
0.1648793533 |
- |
- |
PREDICTED: uncharacterized protein LOC105633681 [Jatropha curcas] |
| 5 |
Hb_002259_060 |
0.1686642852 |
- |
- |
PREDICTED: kinesin-like calmodulin-binding protein isoform X4 [Jatropha curcas] |
| 6 |
Hb_022250_150 |
0.1741872436 |
- |
- |
PREDICTED: uncharacterized protein LOC105648687 [Jatropha curcas] |
| 7 |
Hb_001799_190 |
0.1746194099 |
- |
- |
PREDICTED: uncharacterized protein LOC105643221 [Jatropha curcas] |
| 8 |
Hb_008053_060 |
0.1854458976 |
- |
- |
PREDICTED: protein PAIR1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_005180_010 |
0.1862313373 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 10 |
Hb_003626_040 |
0.1890593473 |
- |
- |
PREDICTED: probable 2-oxoglutarate-dependent dioxygenase At3g49630 isoform X2 [Jatropha curcas] |
| 11 |
Hb_002836_100 |
0.1895376411 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000395_220 |
0.190195042 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 13 |
Hb_005489_140 |
0.1913096613 |
- |
- |
PREDICTED: maspardin [Jatropha curcas] |
| 14 |
Hb_001227_100 |
0.1920341646 |
- |
- |
RecName: Full=Enolase 1; AltName: Full=2-phospho-D-glycerate hydro-lyase 1; AltName: Full=2-phosphoglycerate dehydratase 1; AltName: Allergen=Hev b 9 [Hevea brasiliensis] |
| 15 |
Hb_000362_110 |
0.1938680751 |
- |
- |
PREDICTED: uncharacterized protein LOC104612097 isoform X2 [Nelumbo nucifera] |
| 16 |
Hb_001123_220 |
0.1966124407 |
- |
- |
- |
| 17 |
Hb_009732_020 |
0.1970287099 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 18 |
Hb_001675_010 |
0.1973393138 |
- |
- |
hypothetical protein TRIUR3_09961 [Triticum urartu] |
| 19 |
Hb_000580_050 |
0.2000881533 |
- |
- |
- |
| 20 |
Hb_025336_020 |
0.2002753627 |
- |
- |
PREDICTED: PP2A regulatory subunit TAP46 [Jatropha curcas] |