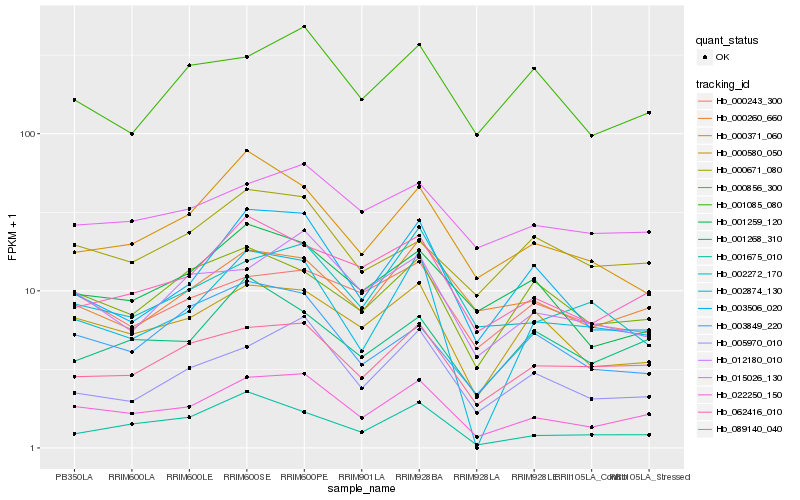
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022250_150 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648687 [Jatropha curcas] |
| 2 |
Hb_089140_040 |
0.1090779041 |
- |
- |
PREDICTED: uncharacterized protein LOC105637966 [Jatropha curcas] |
| 3 |
Hb_000580_050 |
0.1100059347 |
- |
- |
- |
| 4 |
Hb_005970_010 |
0.1141691124 |
- |
- |
PREDICTED: probable isoprenylcysteine alpha-carbonyl methylesterase ICMEL1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_062416_010 |
0.1180086036 |
- |
- |
PREDICTED: UPF0202 protein At1g10490 [Jatropha curcas] |
| 6 |
Hb_003506_020 |
0.1196230611 |
- |
- |
PREDICTED: uncharacterized protein LOC105125881 isoform X1 [Populus euphratica] |
| 7 |
Hb_012180_010 |
0.1204694814 |
- |
- |
PREDICTED: (S)-ureidoglycine aminohydrolase [Jatropha curcas] |
| 8 |
Hb_000260_660 |
0.1272257301 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000856_300 |
0.1289982539 |
- |
- |
PREDICTED: uncharacterized protein LOC105640498 [Jatropha curcas] |
| 10 |
Hb_002874_130 |
0.1292631625 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 9 [Jatropha curcas] |
| 11 |
Hb_015026_130 |
0.1299741554 |
- |
- |
quinone oxidoreductase, putative [Ricinus communis] |
| 12 |
Hb_001259_120 |
0.130859051 |
- |
- |
hypothetical protein EUGRSUZ_C043222, partial [Eucalyptus grandis] |
| 13 |
Hb_001085_080 |
0.1334739041 |
- |
- |
eukaryotic translation elongation factor 1-alpha [Hevea brasiliensis] |
| 14 |
Hb_000371_060 |
0.1357196203 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 15 |
Hb_003849_220 |
0.1358689261 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Jatropha curcas] |
| 16 |
Hb_000243_300 |
0.1367726108 |
- |
- |
catalytic, putative [Ricinus communis] |
| 17 |
Hb_000671_080 |
0.1375990289 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 4 [Jatropha curcas] |
| 18 |
Hb_001675_010 |
0.1378350169 |
- |
- |
hypothetical protein TRIUR3_09961 [Triticum urartu] |
| 19 |
Hb_001268_310 |
0.1381329725 |
- |
- |
- |
| 20 |
Hb_002272_170 |
0.138762916 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase-like protein 1 [Jatropha curcas] |