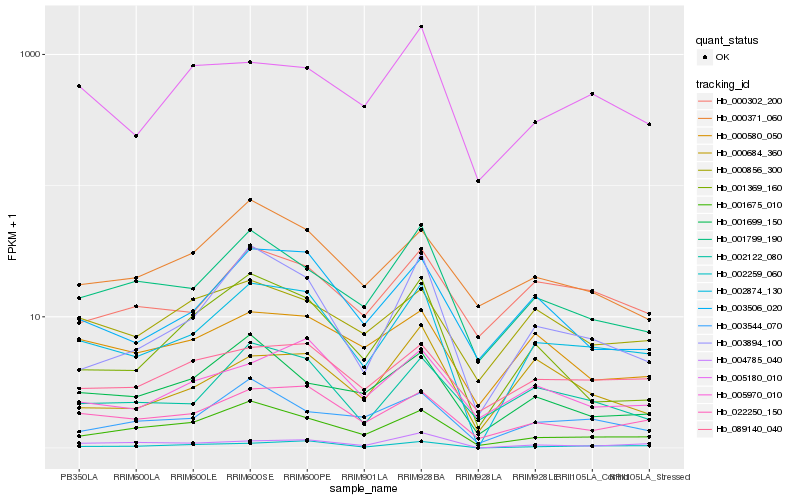
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002874_130 |
0.0 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 9 [Jatropha curcas] |
| 2 |
Hb_002259_060 |
0.1209561351 |
- |
- |
PREDICTED: kinesin-like calmodulin-binding protein isoform X4 [Jatropha curcas] |
| 3 |
Hb_022250_150 |
0.1292631625 |
- |
- |
PREDICTED: uncharacterized protein LOC105648687 [Jatropha curcas] |
| 4 |
Hb_001675_010 |
0.1413623824 |
- |
- |
hypothetical protein TRIUR3_09961 [Triticum urartu] |
| 5 |
Hb_003894_100 |
0.1466507291 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1-like [Jatropha curcas] |
| 6 |
Hb_003506_020 |
0.1488964618 |
- |
- |
PREDICTED: uncharacterized protein LOC105125881 isoform X1 [Populus euphratica] |
| 7 |
Hb_002122_080 |
0.1515326846 |
- |
- |
- |
| 8 |
Hb_001799_190 |
0.1529054956 |
- |
- |
PREDICTED: uncharacterized protein LOC105643221 [Jatropha curcas] |
| 9 |
Hb_089140_040 |
0.1592946856 |
- |
- |
PREDICTED: uncharacterized protein LOC105637966 [Jatropha curcas] |
| 10 |
Hb_004785_040 |
0.15957265 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 3-like [Jatropha curcas] |
| 11 |
Hb_003544_070 |
0.1635682622 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005970_010 |
0.1643225351 |
- |
- |
PREDICTED: probable isoprenylcysteine alpha-carbonyl methylesterase ICMEL1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000580_050 |
0.1661024729 |
- |
- |
- |
| 14 |
Hb_001699_150 |
0.1692943564 |
- |
- |
hypothetical protein POPTR_0009s13450g [Populus trichocarpa] |
| 15 |
Hb_000302_200 |
0.1704483608 |
- |
- |
ornithine cyclodeaminase, putative [Ricinus communis] |
| 16 |
Hb_005180_010 |
0.170865211 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 17 |
Hb_001369_160 |
0.1708725952 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 18 |
Hb_000856_300 |
0.1712875496 |
- |
- |
PREDICTED: uncharacterized protein LOC105640498 [Jatropha curcas] |
| 19 |
Hb_000371_060 |
0.1736661669 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 20 |
Hb_000684_360 |
0.1739994722 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_16454 [Jatropha curcas] |