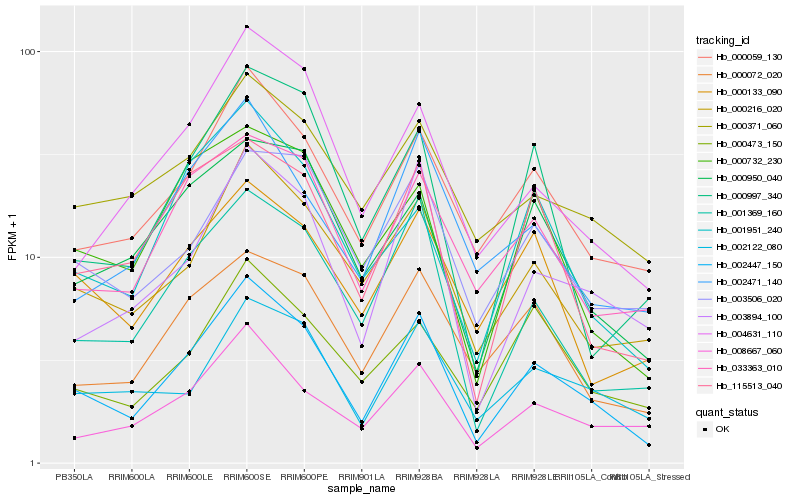
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002447_150 |
0.0 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 2 |
Hb_000059_130 |
0.1181499556 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 3 |
Hb_000216_020 |
0.1200612763 |
- |
- |
PREDICTED: benzaldehyde dehydrogenase (NAD(+))-like [Jatropha curcas] |
| 4 |
Hb_001369_160 |
0.1234825644 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 5 |
Hb_003894_100 |
0.1268834521 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1-like [Jatropha curcas] |
| 6 |
Hb_008667_060 |
0.1343627703 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 7 |
Hb_115513_040 |
0.1346073377 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000072_020 |
0.1367886512 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 9 |
Hb_033363_010 |
0.1383708555 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 10 |
Hb_002122_080 |
0.1404588984 |
- |
- |
- |
| 11 |
Hb_001951_240 |
0.1421220532 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |
| 12 |
Hb_000950_040 |
0.14234501 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 13 |
Hb_000732_230 |
0.1427188403 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_002471_140 |
0.1439281897 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 15 |
Hb_004631_110 |
0.1444540768 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 16 |
Hb_003506_020 |
0.1460344199 |
- |
- |
PREDICTED: uncharacterized protein LOC105125881 isoform X1 [Populus euphratica] |
| 17 |
Hb_000133_090 |
0.1471638293 |
- |
- |
PREDICTED: uncharacterized protein LOC105629320 [Jatropha curcas] |
| 18 |
Hb_000371_060 |
0.1475604276 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 19 |
Hb_000997_340 |
0.1483482493 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 20 |
Hb_000473_150 |
0.1486023567 |
- |
- |
PREDICTED: pullulanase 1, chloroplastic [Jatropha curcas] |