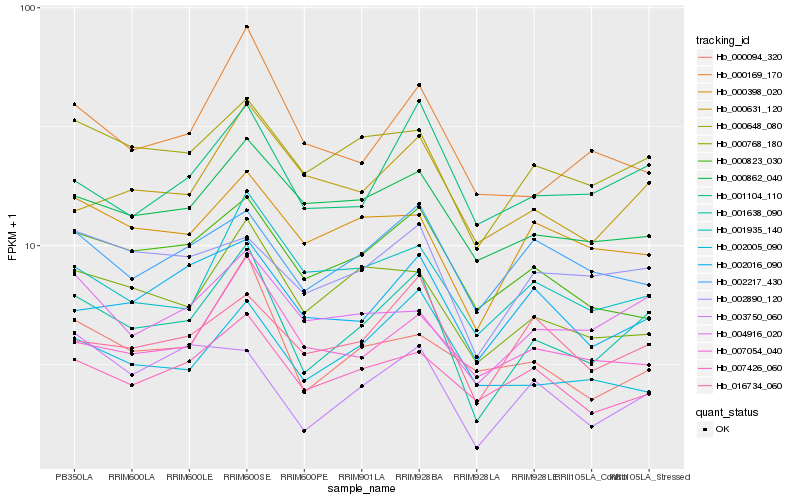
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001638_090 |
0.0 |
- |
- |
GRF1-INTERACTING FACTOR 2 family protein [Populus trichocarpa] |
| 2 |
Hb_000648_080 |
0.110693421 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 1 isoform X2 [Populus euphratica] |
| 3 |
Hb_001104_110 |
0.1207396698 |
- |
- |
PREDICTED: splicing factor U2af small subunit B-like [Populus euphratica] |
| 4 |
Hb_016734_060 |
0.1213450194 |
- |
- |
PREDICTED: nucleolar protein 6 [Jatropha curcas] |
| 5 |
Hb_000768_180 |
0.1214082521 |
- |
- |
PREDICTED: uncharacterized protein LOC105632528 [Jatropha curcas] |
| 6 |
Hb_004916_020 |
0.1236116207 |
- |
- |
PREDICTED: DNA ligase 4 [Jatropha curcas] |
| 7 |
Hb_002890_120 |
0.1255914864 |
- |
- |
Nucleic acid binding,RNA binding isoform 1 [Theobroma cacao] |
| 8 |
Hb_007426_060 |
0.1262865035 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 9 |
Hb_000398_020 |
0.1263804413 |
- |
- |
PREDICTED: superkiller viralicidic activity 2-like 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002016_090 |
0.1294552457 |
- |
- |
PREDICTED: DNA mismatch repair protein PMS1 [Jatropha curcas] |
| 11 |
Hb_000169_170 |
0.1299259296 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 12 |
Hb_001935_140 |
0.1300675423 |
- |
- |
CCR4-NOT transcription complex subunit 1 [Gossypium arboreum] |
| 13 |
Hb_000862_040 |
0.1303458892 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 14 |
Hb_002217_430 |
0.1309850345 |
- |
- |
PREDICTED: flowering time control protein FCA isoform X4 [Jatropha curcas] |
| 15 |
Hb_000823_030 |
0.1316392859 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC5 [Jatropha curcas] |
| 16 |
Hb_000631_120 |
0.1325192869 |
- |
- |
hypothetical protein CISIN_1g0431902mg, partial [Citrus sinensis] |
| 17 |
Hb_000094_320 |
0.132815957 |
- |
- |
hypothetical protein JCGZ_04407 [Jatropha curcas] |
| 18 |
Hb_007054_040 |
0.1343389242 |
- |
- |
PREDICTED: protein RST1 [Jatropha curcas] |
| 19 |
Hb_003750_060 |
0.1348598561 |
- |
- |
PREDICTED: protein TIC 40, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002005_090 |
0.1356176377 |
- |
- |
conserved hypothetical protein [Ricinus communis] |