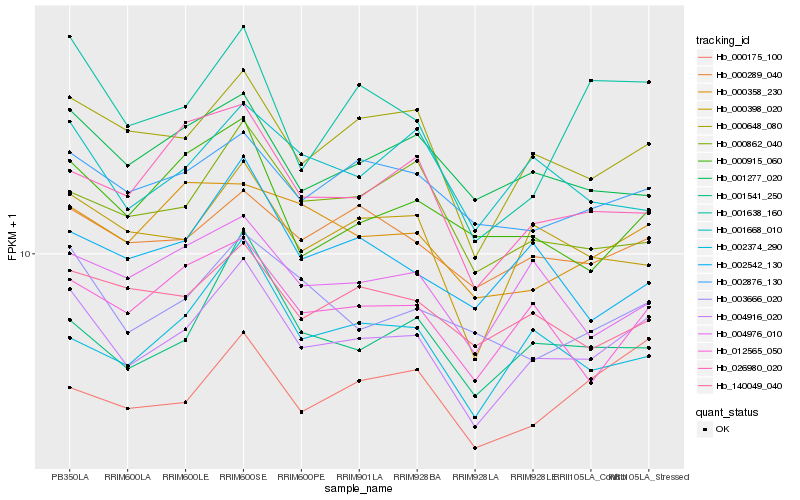
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004916_020 |
0.0 |
- |
- |
PREDICTED: DNA ligase 4 [Jatropha curcas] |
| 2 |
Hb_000648_080 |
0.0744776879 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 1 isoform X2 [Populus euphratica] |
| 3 |
Hb_000398_020 |
0.0863339415 |
- |
- |
PREDICTED: superkiller viralicidic activity 2-like 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001541_250 |
0.0873118204 |
- |
- |
PREDICTED: integrator complex subunit 9 homolog isoform X1 [Jatropha curcas] |
| 5 |
Hb_012565_050 |
0.0904179927 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein 70 kDa [Jatropha curcas] |
| 6 |
Hb_003666_020 |
0.0934568879 |
- |
- |
kinase, putative [Ricinus communis] |
| 7 |
Hb_000289_040 |
0.0944693475 |
- |
- |
Poly(A) polymerase alpha, putative [Ricinus communis] |
| 8 |
Hb_002876_130 |
0.0956719132 |
- |
- |
PREDICTED: uncharacterized protein LOC105637143 [Jatropha curcas] |
| 9 |
Hb_001638_160 |
0.0963409912 |
- |
- |
PREDICTED: uncharacterized protein LOC105642785 [Jatropha curcas] |
| 10 |
Hb_002374_290 |
0.0970685692 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 26 isoform X1 [Jatropha curcas] |
| 11 |
Hb_140049_040 |
0.0986220147 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 12 |
Hb_004976_010 |
0.1012348786 |
- |
- |
hypothetical protein RCOM_1429110 [Ricinus communis] |
| 13 |
Hb_001277_020 |
0.1014917197 |
- |
- |
PREDICTED: armadillo repeat-containing protein LFR [Jatropha curcas] |
| 14 |
Hb_026980_020 |
0.10332667 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0234100-like [Jatropha curcas] |
| 15 |
Hb_000862_040 |
0.1040009115 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 16 |
Hb_001668_010 |
0.1040729643 |
- |
- |
PREDICTED: RNA-binding protein with multiple splicing [Jatropha curcas] |
| 17 |
Hb_000175_100 |
0.1043299147 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 18 |
Hb_000358_230 |
0.1044545377 |
- |
- |
protein phosphatases pp1 regulatory subunit, putative [Ricinus communis] |
| 19 |
Hb_000915_060 |
0.1050237261 |
- |
- |
PREDICTED: uncharacterized protein LOC105628864 [Jatropha curcas] |
| 20 |
Hb_002542_130 |
0.1056497764 |
transcription factor |
TF Family: HB |
Homeobox protein LUMINIDEPENDENS, putative [Ricinus communis] |