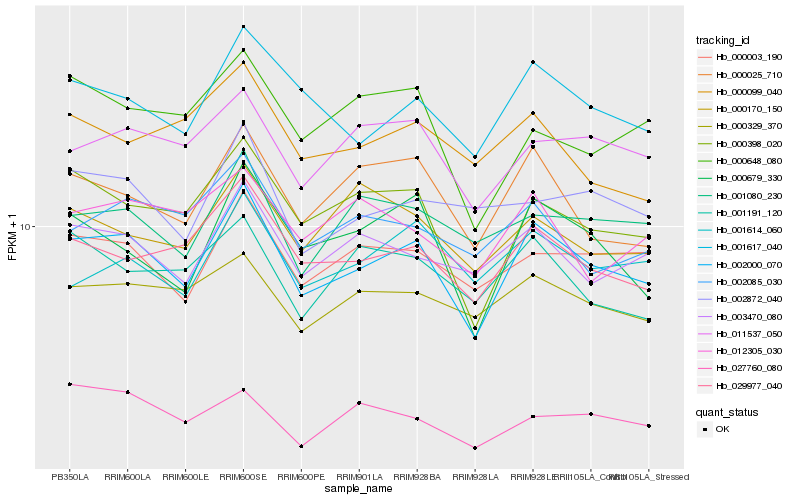
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002000_070 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: 5-formyltetrahydrofolate cyclo-ligase-like protein COG0212 [Jatropha curcas] |
| 2 |
Hb_029977_040 |
0.0734753013 |
- |
- |
PREDICTED: uncharacterized protein LOC105640155 isoform X4 [Jatropha curcas] |
| 3 |
Hb_001617_040 |
0.0749682259 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_000398_020 |
0.0753651821 |
- |
- |
PREDICTED: superkiller viralicidic activity 2-like 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000025_710 |
0.0799889657 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 6 |
Hb_000679_330 |
0.0801241758 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase KEG [Jatropha curcas] |
| 7 |
Hb_000003_190 |
0.0806888967 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA-like isoform X1 [Nelumbo nucifera] |
| 8 |
Hb_011537_050 |
0.0825623188 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003470_080 |
0.0839637766 |
transcription factor |
TF Family: SNF2 |
PREDICTED: protein PHOTOPERIOD-INDEPENDENT EARLY FLOWERING 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000329_370 |
0.0888959543 |
- |
- |
hypothetical protein JCGZ_14571 [Jatropha curcas] |
| 11 |
Hb_001080_230 |
0.0899860448 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105649979 [Jatropha curcas] |
| 12 |
Hb_001614_060 |
0.0919235653 |
- |
- |
hypothetical protein JCGZ_06918 [Jatropha curcas] |
| 13 |
Hb_002085_030 |
0.0928581564 |
- |
- |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 14 |
Hb_002872_040 |
0.0940202915 |
- |
- |
pumilio, putative [Ricinus communis] |
| 15 |
Hb_001191_120 |
0.0947919372 |
- |
- |
PREDICTED: cleavage stimulating factor 64 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000648_080 |
0.0960992283 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 1 isoform X2 [Populus euphratica] |
| 17 |
Hb_000170_150 |
0.0964802911 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 18 |
Hb_027760_080 |
0.0985429626 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_012305_030 |
0.0993531209 |
- |
- |
hypothetical protein JCGZ_18979 [Jatropha curcas] |
| 20 |
Hb_000099_040 |
0.0995275295 |
- |
- |
unnamed protein product [Coffea canephora] |