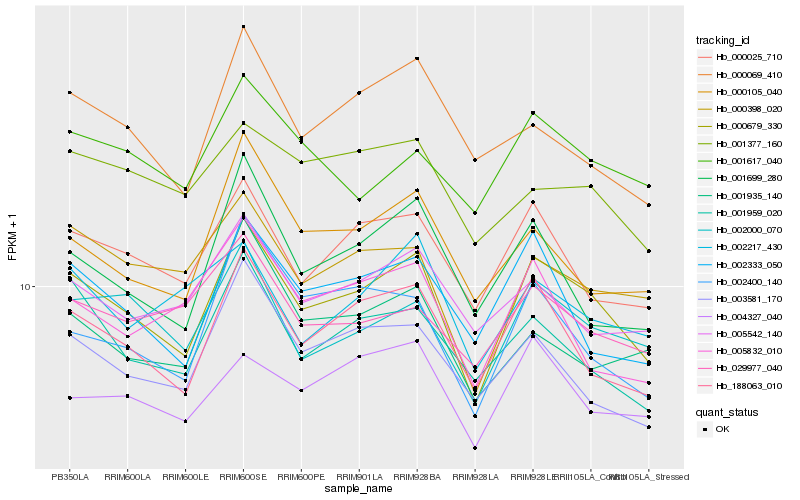
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000679_330 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase KEG [Jatropha curcas] |
| 2 |
Hb_188063_010 |
0.0702261049 |
- |
- |
hypothetical protein RCOM_1469330 [Ricinus communis] |
| 3 |
Hb_002000_070 |
0.0801241758 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: 5-formyltetrahydrofolate cyclo-ligase-like protein COG0212 [Jatropha curcas] |
| 4 |
Hb_000025_710 |
0.0805002436 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 5 |
Hb_002333_050 |
0.0862653799 |
- |
- |
PREDICTED: uncharacterized protein LOC105644854 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000398_020 |
0.0887467578 |
- |
- |
PREDICTED: superkiller viralicidic activity 2-like 2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001959_020 |
0.0898579057 |
- |
- |
PREDICTED: uncharacterized protein LOC105638454 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003581_170 |
0.091376549 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001699_280 |
0.0925451393 |
- |
- |
PREDICTED: transformation/transcription domain-associated protein [Jatropha curcas] |
| 10 |
Hb_002400_140 |
0.0936289836 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 11 |
Hb_000105_040 |
0.0946557733 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOR isoform X2 [Jatropha curcas] |
| 12 |
Hb_005542_140 |
0.0970289109 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 13 |
Hb_004327_040 |
0.0970800001 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 27 homolog [Jatropha curcas] |
| 14 |
Hb_000069_410 |
0.097525548 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 15 |
Hb_001617_040 |
0.0983935403 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 16 |
Hb_005832_010 |
0.1002536622 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 17 |
Hb_029977_040 |
0.1002624676 |
- |
- |
PREDICTED: uncharacterized protein LOC105640155 isoform X4 [Jatropha curcas] |
| 18 |
Hb_001377_160 |
0.1008454074 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 19 |
Hb_001935_140 |
0.1022289918 |
- |
- |
CCR4-NOT transcription complex subunit 1 [Gossypium arboreum] |
| 20 |
Hb_002217_430 |
0.1028916368 |
- |
- |
PREDICTED: flowering time control protein FCA isoform X4 [Jatropha curcas] |