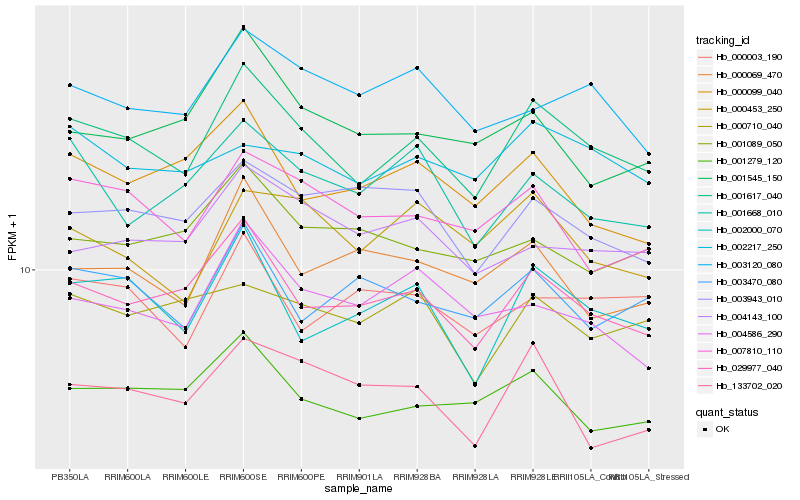
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001617_040 |
0.0 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 2 |
Hb_029977_040 |
0.0637893648 |
- |
- |
PREDICTED: uncharacterized protein LOC105640155 isoform X4 [Jatropha curcas] |
| 3 |
Hb_002000_070 |
0.0749682259 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: 5-formyltetrahydrofolate cyclo-ligase-like protein COG0212 [Jatropha curcas] |
| 4 |
Hb_001279_120 |
0.0785581856 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5-like [Jatropha curcas] |
| 5 |
Hb_007810_110 |
0.0820183845 |
- |
- |
PREDICTED: DNA/RNA-binding protein KIN17 [Jatropha curcas] |
| 6 |
Hb_003120_080 |
0.0900891755 |
- |
- |
PREDICTED: COPII coat assembly protein SEC16 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002217_250 |
0.0909037991 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000453_250 |
0.0915672746 |
- |
- |
PREDICTED: uncharacterized protein LOC105637185 [Jatropha curcas] |
| 9 |
Hb_003470_080 |
0.0917002243 |
transcription factor |
TF Family: SNF2 |
PREDICTED: protein PHOTOPERIOD-INDEPENDENT EARLY FLOWERING 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001668_010 |
0.0920535752 |
- |
- |
PREDICTED: RNA-binding protein with multiple splicing [Jatropha curcas] |
| 11 |
Hb_000099_040 |
0.0923363452 |
- |
- |
unnamed protein product [Coffea canephora] |
| 12 |
Hb_000003_190 |
0.0928264202 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA-like isoform X1 [Nelumbo nucifera] |
| 13 |
Hb_004143_100 |
0.0930045904 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004586_290 |
0.0935326492 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 15 |
Hb_003943_010 |
0.0945409098 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000710_040 |
0.0949519018 |
- |
- |
PREDICTED: HEAT repeat-containing protein 6 [Jatropha curcas] |
| 17 |
Hb_000069_470 |
0.095278086 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001089_050 |
0.0964099006 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 4 [Jatropha curcas] |
| 19 |
Hb_133702_020 |
0.0966917876 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_001545_150 |
0.0967304142 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105635527 [Jatropha curcas] |