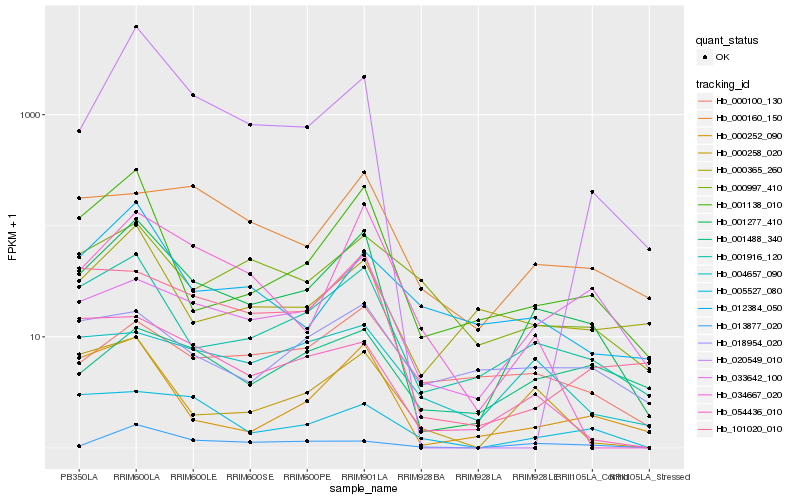
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001277_410 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632036 [Jatropha curcas] |
| 2 |
Hb_001916_120 |
0.154871229 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 3 |
Hb_001138_010 |
0.19453516 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_000252_090 |
0.2066987293 |
- |
- |
PREDICTED: L-tryptophan--pyruvate aminotransferase 1 [Jatropha curcas] |
| 5 |
Hb_000258_020 |
0.2165860579 |
- |
- |
PREDICTED: RING-H2 finger protein ATL74 [Jatropha curcas] |
| 6 |
Hb_101020_010 |
0.2268420953 |
- |
- |
- |
| 7 |
Hb_020549_010 |
0.2332660402 |
- |
- |
PREDICTED: uncharacterized protein LOC105638664 isoform X2 [Jatropha curcas] |
| 8 |
Hb_034667_020 |
0.2342791675 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_033642_100 |
0.2372080019 |
- |
- |
PREDICTED: receptor homology region, transmembrane domain- and RING domain-containing protein 1 [Prunus mume] |
| 10 |
Hb_000160_150 |
0.2380622295 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 3, chloroplastic [Cucumis melo] |
| 11 |
Hb_000365_260 |
0.2389105177 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_054436_010 |
0.2419476608 |
- |
- |
PREDICTED: uncharacterized protein LOC105638666 [Jatropha curcas] |
| 13 |
Hb_004657_090 |
0.2421693994 |
- |
- |
Chromosome-associated kinesin KLP1, putative [Ricinus communis] |
| 14 |
Hb_000100_130 |
0.2441797649 |
- |
- |
hypothetical protein VITISV_026702 [Vitis vinifera] |
| 15 |
Hb_013877_020 |
0.2447442137 |
- |
- |
Cytochrome P450 [Theobroma cacao] |
| 16 |
Hb_005527_080 |
0.2457229415 |
- |
- |
hypothetical protein CICLE_v10023951mg, partial [Citrus clementina] |
| 17 |
Hb_018954_020 |
0.2459721452 |
- |
- |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH13 [Jatropha curcas] |
| 18 |
Hb_001488_340 |
0.2479635321 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 19 |
Hb_000997_410 |
0.2495200453 |
- |
- |
PREDICTED: uncharacterized protein LOC105632066 [Jatropha curcas] |
| 20 |
Hb_012384_050 |
0.2500585619 |
- |
- |
PREDICTED: uncharacterized protein LOC105641396 [Jatropha curcas] |