| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
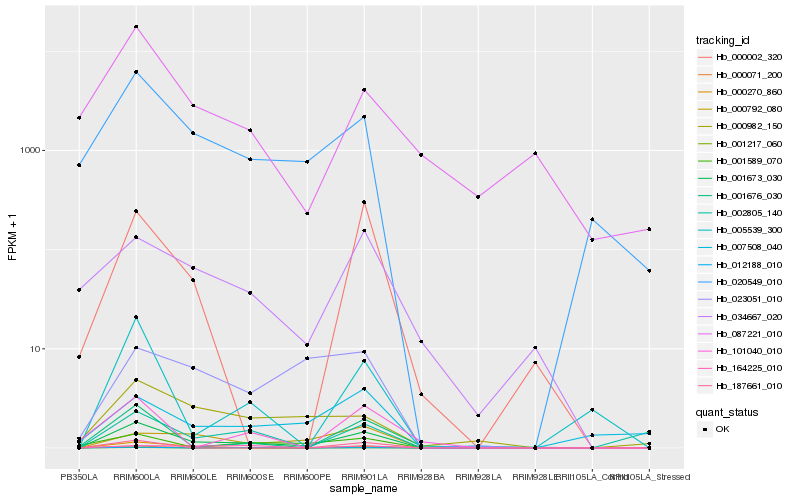
Hb_001673_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_08729 [Jatropha curcas] |
| 2 |
Hb_020549_010 |
0.2317550681 |
- |
- |
PREDICTED: uncharacterized protein LOC105638664 isoform X2 [Jatropha curcas] |
| 3 |
Hb_001676_030 |
0.2625462119 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 4 |
Hb_000071_200 |
0.2636229675 |
- |
- |
PREDICTED: uncharacterized protein LOC105764710 isoform X3 [Gossypium raimondii] |
| 5 |
Hb_000270_860 |
0.267512382 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_005539_300 |
0.2757594443 |
- |
- |
PREDICTED: deacetoxyvindoline 4-hydroxylase-like [Populus euphratica] |
| 7 |
Hb_000792_080 |
0.2864519666 |
- |
- |
hypothetical protein B456_012G111300, partial [Gossypium raimondii] |
| 8 |
Hb_001589_070 |
0.2864873499 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 9 |
Hb_000982_150 |
0.293025005 |
transcription factor |
TF Family: Orphans |
histidine kinase 1 plant, putative [Ricinus communis] |
| 10 |
Hb_023051_010 |
0.3048222962 |
- |
- |
PREDICTED: putative amidase C869.01 [Populus euphratica] |
| 11 |
Hb_000002_320 |
0.305091051 |
- |
- |
PREDICTED: uncharacterized protein LOC105632108 [Jatropha curcas] |
| 12 |
Hb_034667_020 |
0.307148125 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001217_060 |
0.3084245555 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 14 |
Hb_012188_010 |
0.3095702615 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 15 |
Hb_007508_040 |
0.315346087 |
- |
- |
PREDICTED: ATP synthase subunit gamma, mitochondrial [Jatropha curcas] |
| 16 |
Hb_002805_140 |
0.3167171251 |
- |
- |
hypothetical protein POPTR_0001s28390g [Populus trichocarpa] |
| 17 |
Hb_101040_010 |
0.3175350491 |
- |
- |
- |
| 18 |
Hb_087221_010 |
0.3201749068 |
- |
- |
hypothetical protein CISIN_1g033698mg [Citrus sinensis] |
| 19 |
Hb_164225_010 |
0.3245466534 |
- |
- |
- |
| 20 |
Hb_187661_010 |
0.3318367883 |
- |
- |
PREDICTED: thylakoidal processing peptidase 1, chloroplastic-like [Jatropha curcas] |