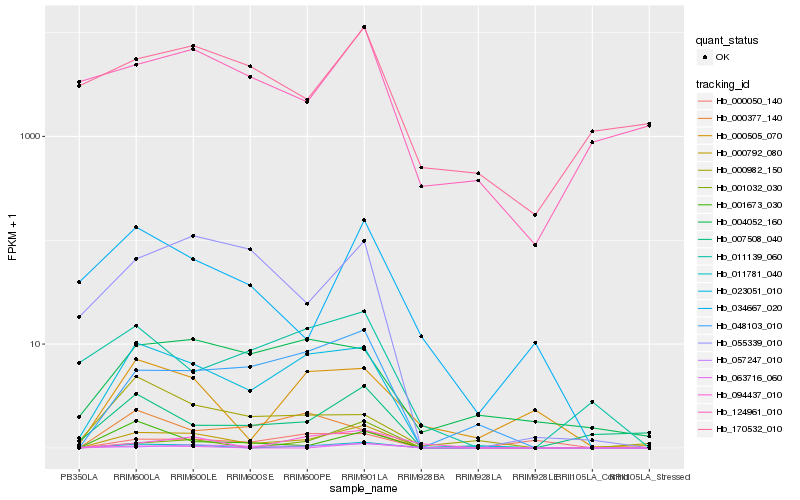
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000792_080 |
0.0 |
- |
- |
hypothetical protein B456_012G111300, partial [Gossypium raimondii] |
| 2 |
Hb_023051_010 |
0.1689383883 |
- |
- |
PREDICTED: putative amidase C869.01 [Populus euphratica] |
| 3 |
Hb_011781_040 |
0.1757168215 |
- |
- |
- |
| 4 |
Hb_048103_010 |
0.229954868 |
- |
- |
- |
| 5 |
Hb_055339_010 |
0.2758708387 |
- |
- |
EARLY-RESPONSIVE TO DEHYDRATION 7 family protein [Populus trichocarpa] |
| 6 |
Hb_001673_030 |
0.2864519666 |
- |
- |
hypothetical protein JCGZ_08729 [Jatropha curcas] |
| 7 |
Hb_001032_030 |
0.2874893863 |
- |
- |
PREDICTED: 60S ribosomal protein L23a-like [Eucalyptus grandis] |
| 8 |
Hb_007508_040 |
0.2925156967 |
- |
- |
PREDICTED: ATP synthase subunit gamma, mitochondrial [Jatropha curcas] |
| 9 |
Hb_000505_070 |
0.3096703314 |
- |
- |
PREDICTED: heparanase-like protein 3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004052_160 |
0.3157780401 |
- |
- |
Chain A, Rna Binding Protein |
| 11 |
Hb_034667_020 |
0.3164848889 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_057247_010 |
0.3212421889 |
- |
- |
hypothetical protein JCGZ_04800 [Jatropha curcas] |
| 13 |
Hb_000982_150 |
0.3216710481 |
transcription factor |
TF Family: Orphans |
histidine kinase 1 plant, putative [Ricinus communis] |
| 14 |
Hb_063716_060 |
0.3228741493 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_094437_010 |
0.3237209251 |
- |
- |
- |
| 16 |
Hb_000050_140 |
0.3249125786 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 2 isoform X1 [Populus euphratica] |
| 17 |
Hb_011139_060 |
0.3317301921 |
- |
- |
- |
| 18 |
Hb_000377_140 |
0.3327157538 |
- |
- |
- |
| 19 |
Hb_124961_010 |
0.3328357298 |
- |
- |
hypothetical protein EUGRSUZ_B03893 [Eucalyptus grandis] |
| 20 |
Hb_170532_010 |
0.3366900826 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 8 [Jatropha curcas] |