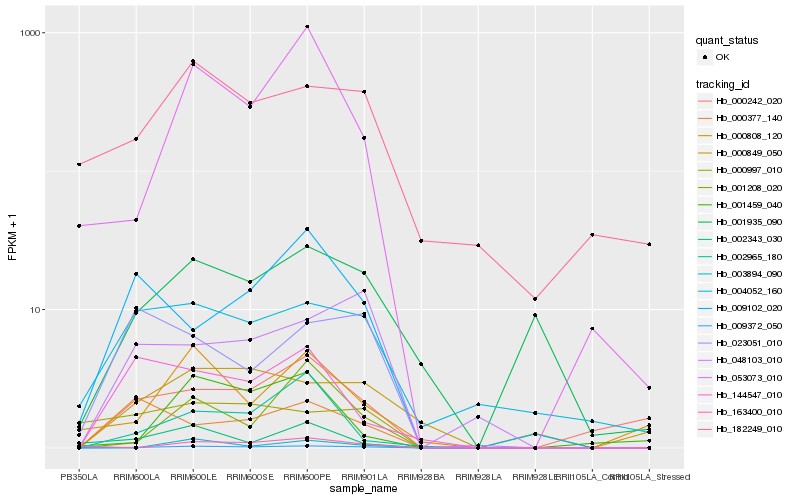
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009372_050 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_053073_010 |
0.2324654555 |
- |
- |
- |
| 3 |
Hb_000242_020 |
0.2494578671 |
- |
- |
hypothetical protein CICLE_v100246891mg, partial [Citrus clementina] |
| 4 |
Hb_001208_020 |
0.2502256799 |
- |
- |
- |
| 5 |
Hb_001935_090 |
0.2514155586 |
- |
- |
PREDICTED: uncharacterized protein LOC105643180 isoform X1 [Jatropha curcas] |
| 6 |
Hb_048103_010 |
0.2557944851 |
- |
- |
- |
| 7 |
Hb_004052_160 |
0.2568987599 |
- |
- |
Chain A, Rna Binding Protein |
| 8 |
Hb_000808_120 |
0.2612521865 |
- |
- |
- |
| 9 |
Hb_000849_050 |
0.2637752197 |
- |
- |
hypothetical protein VITISV_010852 [Vitis vinifera] |
| 10 |
Hb_182249_010 |
0.2642122022 |
- |
- |
- |
| 11 |
Hb_002965_180 |
0.2656368254 |
- |
- |
AMSH-like ubiquitin thioesterase 3 [Morus notabilis] |
| 12 |
Hb_144547_010 |
0.2710068437 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 13 |
Hb_009102_020 |
0.271036765 |
- |
- |
hypothetical protein EUGRSUZ_I01262 [Eucalyptus grandis] |
| 14 |
Hb_003894_090 |
0.2749008504 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002343_030 |
0.2778292708 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000997_010 |
0.2798203752 |
- |
- |
PREDICTED: FHA domain-containing protein At4g14490 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000377_140 |
0.2801996954 |
- |
- |
- |
| 18 |
Hb_023051_010 |
0.2820067569 |
- |
- |
PREDICTED: putative amidase C869.01 [Populus euphratica] |
| 19 |
Hb_001459_040 |
0.284378025 |
- |
- |
Ankyrin repeat-containing protein [Medicago truncatula] |
| 20 |
Hb_163400_010 |
0.2869971993 |
- |
- |
PREDICTED: uncharacterized protein LOC105631661 [Jatropha curcas] |