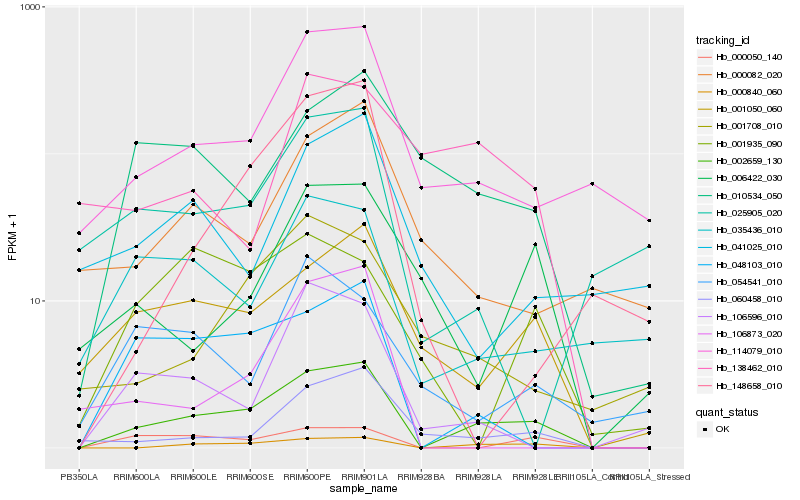
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002659_130 |
0.0 |
- |
- |
- |
| 2 |
Hb_000840_060 |
0.1843171153 |
- |
- |
- |
| 3 |
Hb_001050_060 |
0.2453977606 |
- |
- |
hypothetical protein POPTR_0011s036602g, partial [Populus trichocarpa] |
| 4 |
Hb_114079_010 |
0.2541964111 |
- |
- |
hypothetical protein CICLE_v10008147mg [Citrus clementina] |
| 5 |
Hb_048103_010 |
0.2557868531 |
- |
- |
- |
| 6 |
Hb_060458_010 |
0.2616468172 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000050_140 |
0.2629051755 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 2 isoform X1 [Populus euphratica] |
| 8 |
Hb_010534_050 |
0.2802880058 |
- |
- |
PREDICTED: 40S ribosomal protein S7 [Jatropha curcas] |
| 9 |
Hb_035436_010 |
0.2897512562 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10-B-like [Populus euphratica] |
| 10 |
Hb_106596_010 |
0.2951108637 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001708_010 |
0.2951633173 |
- |
- |
PREDICTED: putative disease resistance protein At4g11170 [Malus domestica] |
| 12 |
Hb_000082_020 |
0.3034364179 |
- |
- |
- |
| 13 |
Hb_006422_030 |
0.306681435 |
- |
- |
- |
| 14 |
Hb_148658_010 |
0.316749362 |
- |
- |
- |
| 15 |
Hb_106873_020 |
0.3168677275 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 16 |
Hb_025905_020 |
0.3180308795 |
- |
- |
major allergen Pru ar 1-like [Jatropha curcas] |
| 17 |
Hb_054541_010 |
0.318254356 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X3 [Jatropha curcas] |
| 18 |
Hb_001935_090 |
0.3200249266 |
- |
- |
PREDICTED: uncharacterized protein LOC105643180 isoform X1 [Jatropha curcas] |
| 19 |
Hb_041025_010 |
0.3223508429 |
- |
- |
PREDICTED: uncharacterized protein LOC105635593 [Jatropha curcas] |
| 20 |
Hb_138462_010 |
0.3223614078 |
- |
- |
PREDICTED: SUMO-conjugating enzyme UBC9-A-like isoform X1 [Elaeis guineensis] |