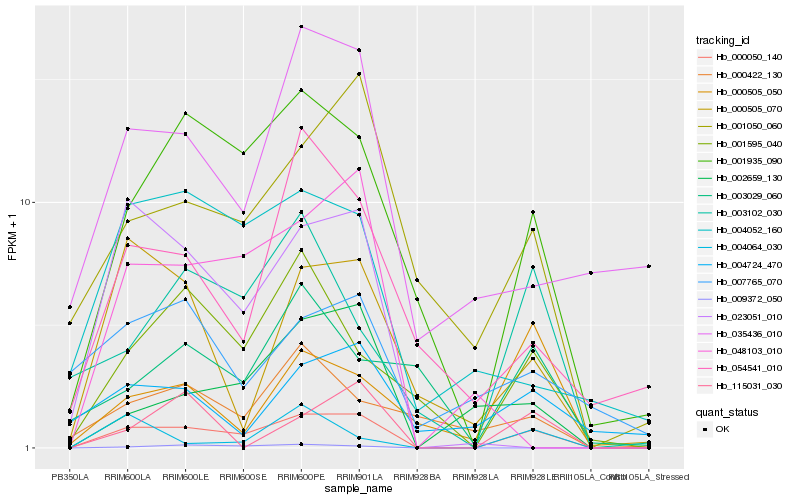
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000050_140 |
0.0 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 2 isoform X1 [Populus euphratica] |
| 2 |
Hb_001935_090 |
0.2114492786 |
- |
- |
PREDICTED: uncharacterized protein LOC105643180 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001595_040 |
0.2628433705 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 4 |
Hb_002659_130 |
0.2629051755 |
- |
- |
- |
| 5 |
Hb_001050_060 |
0.2630040265 |
- |
- |
hypothetical protein POPTR_0011s036602g, partial [Populus trichocarpa] |
| 6 |
Hb_000505_070 |
0.2848511954 |
- |
- |
PREDICTED: heparanase-like protein 3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003102_030 |
0.2865095176 |
- |
- |
PREDICTED: uncharacterized protein LOC105644418 [Jatropha curcas] |
| 8 |
Hb_048103_010 |
0.2910437478 |
- |
- |
- |
| 9 |
Hb_023051_010 |
0.2917366442 |
- |
- |
PREDICTED: putative amidase C869.01 [Populus euphratica] |
| 10 |
Hb_004052_160 |
0.2942278942 |
- |
- |
Chain A, Rna Binding Protein |
| 11 |
Hb_054541_010 |
0.2947198221 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X3 [Jatropha curcas] |
| 12 |
Hb_035436_010 |
0.2981881666 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10-B-like [Populus euphratica] |
| 13 |
Hb_115031_030 |
0.2992221369 |
- |
- |
ribosomal protein S12 (chloroplast) [Camellia sinensis] |
| 14 |
Hb_000505_050 |
0.3094164372 |
- |
- |
PREDICTED: heparanase-like protein 3 isoform X2 [Gossypium raimondii] |
| 15 |
Hb_009372_050 |
0.3129543789 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_000422_130 |
0.3137754595 |
- |
- |
hypothetical protein JCGZ_15217 [Jatropha curcas] |
| 17 |
Hb_004724_470 |
0.3146314924 |
- |
- |
Nitrilase, putative [Ricinus communis] |
| 18 |
Hb_004064_030 |
0.3156301308 |
- |
- |
aldose-1-epimerase, putative [Ricinus communis] |
| 19 |
Hb_003029_060 |
0.3204429601 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_007765_070 |
0.3205759162 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 2 [Jatropha curcas] |