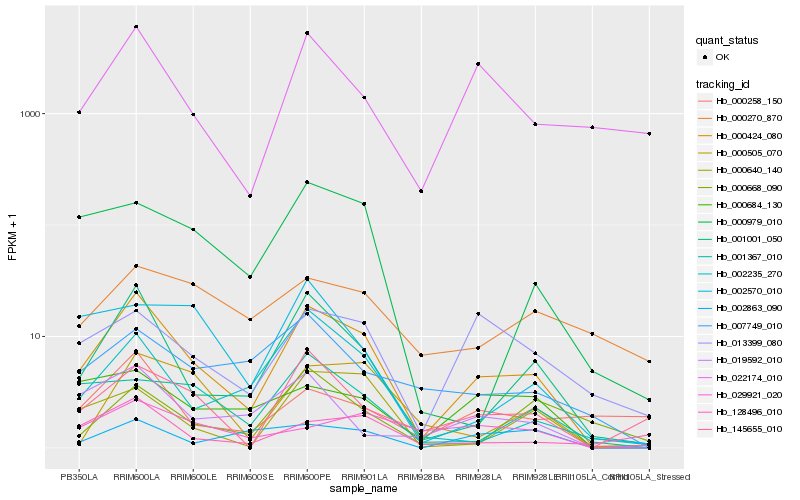
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000424_080 |
0.0 |
- |
- |
PREDICTED: endoglucanase 16 [Jatropha curcas] |
| 2 |
Hb_001001_050 |
0.1694358139 |
- |
- |
CXE protein [Hevea brasiliensis] |
| 3 |
Hb_145655_010 |
0.244118764 |
- |
- |
PREDICTED: lactoylglutathione lyase [Jatropha curcas] |
| 4 |
Hb_022174_010 |
0.2487990415 |
- |
- |
elicitor-responsive protein [Hevea brasiliensis] |
| 5 |
Hb_002235_270 |
0.2512905386 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 100 [Jatropha curcas] |
| 6 |
Hb_000640_140 |
0.2546496766 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000979_010 |
0.2552018238 |
- |
- |
PREDICTED: uncharacterized protein LOC105128288 [Populus euphratica] |
| 8 |
Hb_013399_080 |
0.258619009 |
- |
- |
PREDICTED: protein Brevis radix-like 2 [Jatropha curcas] |
| 9 |
Hb_019592_010 |
0.2592958091 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Pyrus x bretschneideri] |
| 10 |
Hb_000505_070 |
0.2612952405 |
- |
- |
PREDICTED: heparanase-like protein 3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001367_010 |
0.2637562679 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 12 |
Hb_128496_010 |
0.2646476764 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_007749_010 |
0.2654158056 |
- |
- |
- |
| 14 |
Hb_000684_130 |
0.2705419146 |
- |
- |
- |
| 15 |
Hb_002863_090 |
0.2771528611 |
desease resistance |
Gene Name: Speriolin_N |
hypothetical protein JCGZ_06191 [Jatropha curcas] |
| 16 |
Hb_002570_010 |
0.2816735363 |
- |
- |
hypothetical protein JCGZ_25329 [Jatropha curcas] |
| 17 |
Hb_000258_150 |
0.2819333666 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 11-like [Jatropha curcas] |
| 18 |
Hb_029921_020 |
0.2874077963 |
- |
- |
PREDICTED: putative transferase CAF17 homolog, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000668_090 |
0.2912274888 |
- |
- |
senescence-associated family protein [Populus trichocarpa] |
| 20 |
Hb_000270_870 |
0.2942475151 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |