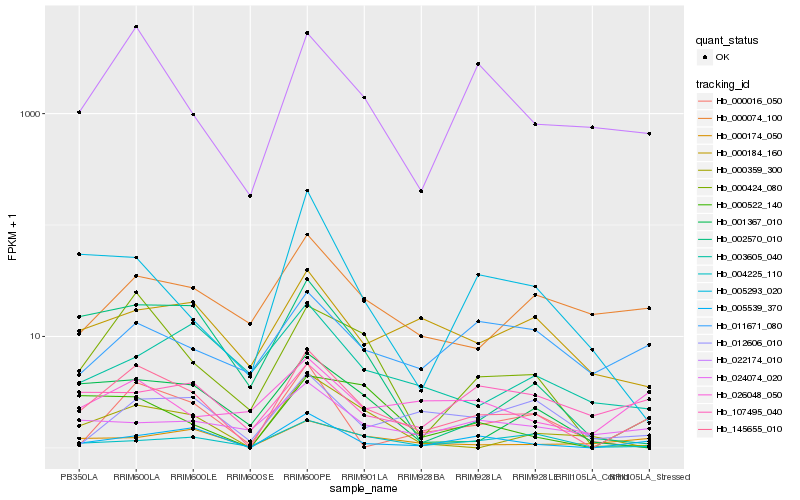
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_145655_010 |
0.0 |
- |
- |
PREDICTED: lactoylglutathione lyase [Jatropha curcas] |
| 2 |
Hb_005539_370 |
0.2076719011 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 3 |
Hb_022174_010 |
0.240959296 |
- |
- |
elicitor-responsive protein [Hevea brasiliensis] |
| 4 |
Hb_000424_080 |
0.244118764 |
- |
- |
PREDICTED: endoglucanase 16 [Jatropha curcas] |
| 5 |
Hb_000174_050 |
0.2666675292 |
- |
- |
PREDICTED: gamma-tubulin complex component 2 [Populus euphratica] |
| 6 |
Hb_000522_140 |
0.2675524837 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 7 |
Hb_004225_110 |
0.2740696738 |
- |
- |
PREDICTED: ABC transporter G family member 28 [Jatropha curcas] |
| 8 |
Hb_000074_100 |
0.2742859649 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000184_160 |
0.2797432022 |
- |
- |
PREDICTED: uncharacterized protein LOC105647991 [Jatropha curcas] |
| 10 |
Hb_002570_010 |
0.2841346211 |
- |
- |
hypothetical protein JCGZ_25329 [Jatropha curcas] |
| 11 |
Hb_001367_010 |
0.2856543071 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 12 |
Hb_000359_300 |
0.2874753527 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At1g06470 [Zea mays] |
| 13 |
Hb_012606_010 |
0.288777932 |
transcription factor |
TF Family: IWS1 |
PREDICTED: probable mediator of RNA polymerase II transcription subunit 26c isoform X2 [Jatropha curcas] |
| 14 |
Hb_024074_020 |
0.2891542544 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g09600 [Jatropha curcas] |
| 15 |
Hb_026048_050 |
0.2910148763 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 2 [Jatropha curcas] |
| 16 |
Hb_011671_080 |
0.2954046314 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase TDR [Jatropha curcas] |
| 17 |
Hb_005293_020 |
0.2964670645 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000016_050 |
0.2964902638 |
- |
- |
PREDICTED: uncharacterized protein LOC105648535 [Jatropha curcas] |
| 19 |
Hb_107495_040 |
0.2967026562 |
- |
- |
PREDICTED: 12-oxophytodienoate reductase 2-like [Citrus sinensis] |
| 20 |
Hb_003605_040 |
0.2981448805 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2 isoform X1 [Jatropha curcas] |