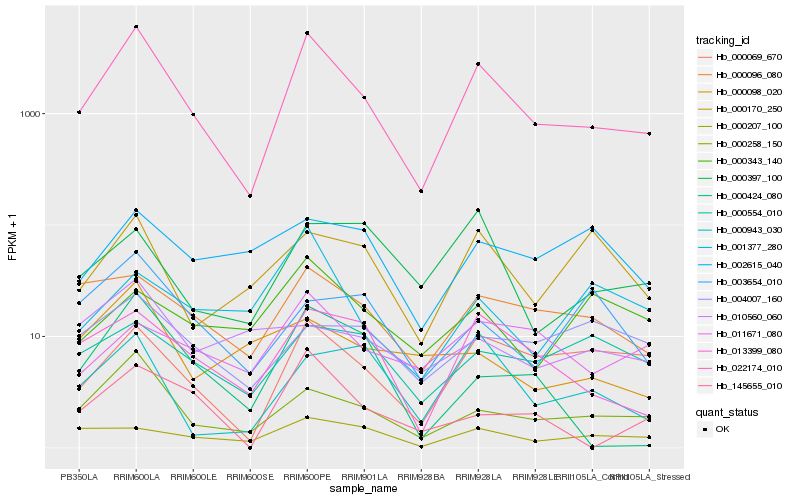
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022174_010 |
0.0 |
- |
- |
elicitor-responsive protein [Hevea brasiliensis] |
| 2 |
Hb_000258_150 |
0.181846249 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 11-like [Jatropha curcas] |
| 3 |
Hb_013399_080 |
0.2019240492 |
- |
- |
PREDICTED: protein Brevis radix-like 2 [Jatropha curcas] |
| 4 |
Hb_000069_670 |
0.2099345147 |
- |
- |
PREDICTED: uncharacterized protein LOC105642887 [Jatropha curcas] |
| 5 |
Hb_000096_080 |
0.2189785255 |
- |
- |
PREDICTED: putative glycosyltransferase 7 [Jatropha curcas] |
| 6 |
Hb_145655_010 |
0.240959296 |
- |
- |
PREDICTED: lactoylglutathione lyase [Jatropha curcas] |
| 7 |
Hb_000207_100 |
0.2466945583 |
- |
- |
PREDICTED: ABC transporter G family member 25 [Jatropha curcas] |
| 8 |
Hb_000943_030 |
0.2480608188 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_011671_080 |
0.2484034396 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase TDR [Jatropha curcas] |
| 10 |
Hb_000424_080 |
0.2487990415 |
- |
- |
PREDICTED: endoglucanase 16 [Jatropha curcas] |
| 11 |
Hb_000170_250 |
0.2532923812 |
- |
- |
hypothetical protein F775_29828 [Aegilops tauschii] |
| 12 |
Hb_000554_010 |
0.2560656792 |
- |
- |
transporter, putative [Ricinus communis] |
| 13 |
Hb_000397_100 |
0.2589504337 |
- |
- |
hypothetical protein 31 [Hevea brasiliensis] |
| 14 |
Hb_000343_140 |
0.2653253622 |
- |
- |
PREDICTED: uncharacterized protein LOC105634966 [Jatropha curcas] |
| 15 |
Hb_000098_020 |
0.2667575864 |
transcription factor |
TF Family: GRAS |
GRAS family transcription factor family protein [Theobroma cacao] |
| 16 |
Hb_001377_280 |
0.2675036805 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Populus euphratica] |
| 17 |
Hb_004007_160 |
0.2677415949 |
- |
- |
PREDICTED: formin-like protein 6 [Jatropha curcas] |
| 18 |
Hb_003654_010 |
0.2698770794 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP650 [Jatropha curcas] |
| 19 |
Hb_002615_040 |
0.2730577599 |
- |
- |
PREDICTED: uncharacterized protein LOC105110770 [Populus euphratica] |
| 20 |
Hb_010560_060 |
0.2772099268 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |