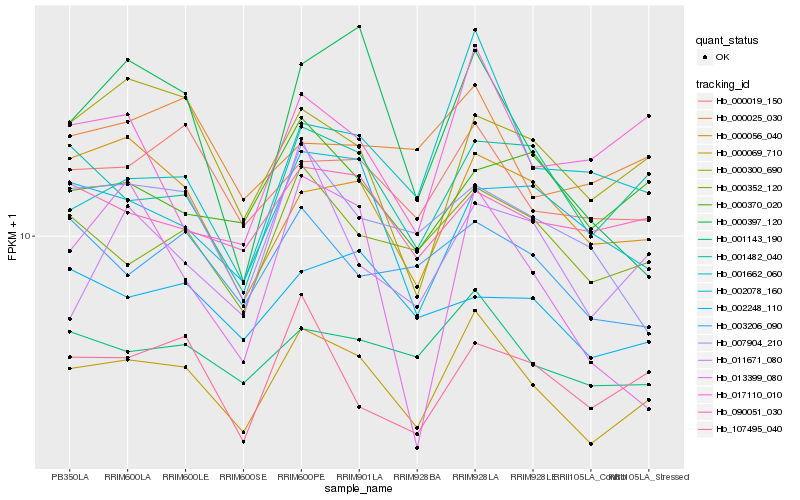
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000069_710 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642890 [Jatropha curcas] |
| 2 |
Hb_001143_190 |
0.1454261318 |
- |
- |
PREDICTED: uncharacterized protein LOC105638976 [Jatropha curcas] |
| 3 |
Hb_000352_120 |
0.1547192813 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105630380 [Jatropha curcas] |
| 4 |
Hb_001482_040 |
0.1560488363 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000019_150 |
0.1586523084 |
- |
- |
PREDICTED: uncharacterized protein LOC105630208 [Jatropha curcas] |
| 6 |
Hb_000056_040 |
0.1605530373 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31 [Jatropha curcas] |
| 7 |
Hb_000397_120 |
0.1626992887 |
- |
- |
PREDICTED: uncharacterized protein LOC105641234 [Jatropha curcas] |
| 8 |
Hb_000300_690 |
0.1677738327 |
- |
- |
hypothetical protein JCGZ_02719 [Jatropha curcas] |
| 9 |
Hb_002078_160 |
0.1707334236 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 10 |
Hb_107495_040 |
0.1748006608 |
- |
- |
PREDICTED: 12-oxophytodienoate reductase 2-like [Citrus sinensis] |
| 11 |
Hb_003206_090 |
0.1759612773 |
- |
- |
PREDICTED: protein trichome birefringence-like 5 [Jatropha curcas] |
| 12 |
Hb_007904_210 |
0.1762316257 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 1D [Jatropha curcas] |
| 13 |
Hb_017110_010 |
0.1765330514 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At1g55760 [Jatropha curcas] |
| 14 |
Hb_090051_030 |
0.1828182307 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 15 |
Hb_001662_060 |
0.1834184034 |
- |
- |
SEC14 cytosolic factor family protein / phosphoglyceride transfer family protein isoform 1 [Theobroma cacao] |
| 16 |
Hb_002248_110 |
0.1868977835 |
- |
- |
hypothetical protein POPTR_0001s41050g [Populus trichocarpa] |
| 17 |
Hb_011671_080 |
0.187356626 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase TDR [Jatropha curcas] |
| 18 |
Hb_000370_020 |
0.1879935774 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_013399_080 |
0.1881179224 |
- |
- |
PREDICTED: protein Brevis radix-like 2 [Jatropha curcas] |
| 20 |
Hb_000025_030 |
0.1883207503 |
- |
- |
transcription factor, putative [Ricinus communis] |