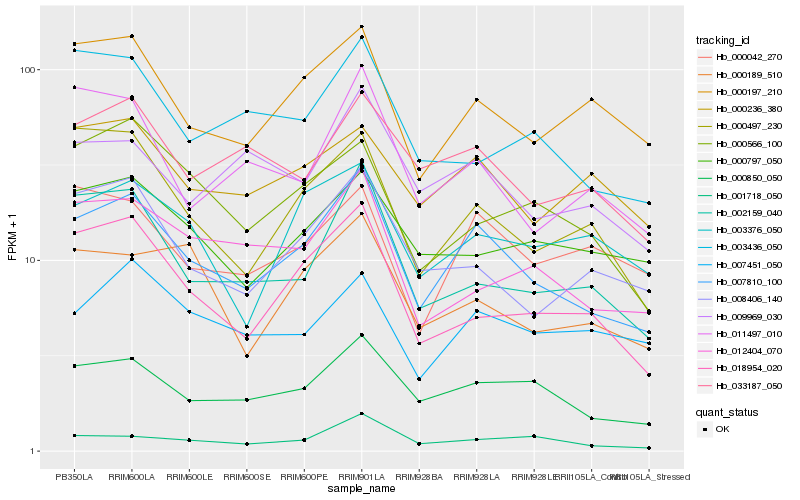
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007810_100 |
0.0 |
- |
- |
Synaptic glycoprotein SC2, putative [Ricinus communis] |
| 2 |
Hb_000850_050 |
0.0965381597 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g63330-like [Jatropha curcas] |
| 3 |
Hb_018954_020 |
0.1234620895 |
- |
- |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH13 [Jatropha curcas] |
| 4 |
Hb_000197_210 |
0.1260049049 |
- |
- |
phosphatidylinositol transporter, putative [Ricinus communis] |
| 5 |
Hb_002159_040 |
0.1296547862 |
- |
- |
hypothetical protein PRUPE_ppa001563mg [Prunus persica] |
| 6 |
Hb_008406_140 |
0.1298484746 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 7 |
Hb_000497_230 |
0.1316747557 |
- |
- |
PREDICTED: probable protein S-acyltransferase 6 [Jatropha curcas] |
| 8 |
Hb_033187_050 |
0.1316765769 |
- |
- |
PREDICTED: exocyst complex component SEC15B [Jatropha curcas] |
| 9 |
Hb_009969_030 |
0.1344876704 |
- |
- |
PREDICTED: argininosuccinate synthase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_012404_070 |
0.1362628085 |
- |
- |
PREDICTED: uncharacterized protein LOC105629465 [Jatropha curcas] |
| 11 |
Hb_001718_050 |
0.1365608835 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Brassica rapa] |
| 12 |
Hb_011497_010 |
0.139095998 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF126-like [Jatropha curcas] |
| 13 |
Hb_000236_380 |
0.1416630286 |
- |
- |
PREDICTED: serine/threonine-protein kinase D6PK [Jatropha curcas] |
| 14 |
Hb_000042_270 |
0.141915861 |
- |
- |
myosin vIII, putative [Ricinus communis] |
| 15 |
Hb_000189_510 |
0.1448148425 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 16 |
Hb_003376_050 |
0.146062995 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 17 |
Hb_000566_100 |
0.1467393335 |
- |
- |
hypothetical protein JCGZ_17993 [Jatropha curcas] |
| 18 |
Hb_000797_050 |
0.1468842878 |
- |
- |
PREDICTED: uncharacterized protein LOC105645916 [Jatropha curcas] |
| 19 |
Hb_007451_050 |
0.1474556065 |
- |
- |
PREDICTED: uncharacterized protein LOC105643409 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003436_050 |
0.147583873 |
- |
- |
arf gtpase-activating protein, putative [Ricinus communis] |