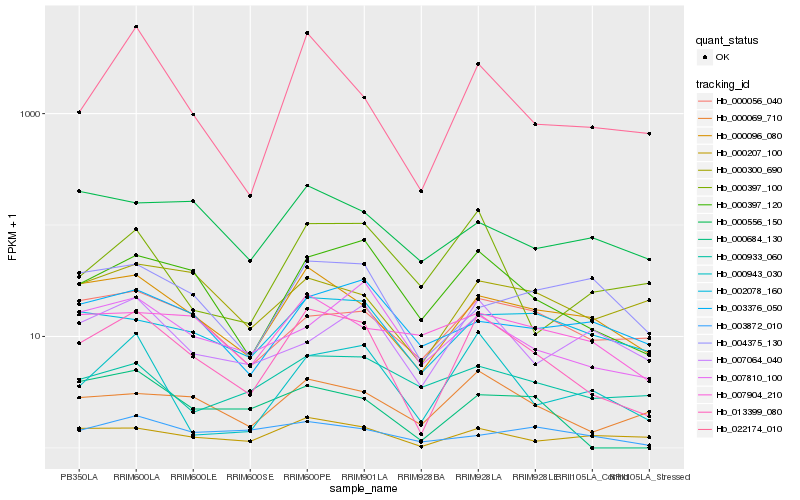
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013399_080 |
0.0 |
- |
- |
PREDICTED: protein Brevis radix-like 2 [Jatropha curcas] |
| 2 |
Hb_000397_120 |
0.1603784579 |
- |
- |
PREDICTED: uncharacterized protein LOC105641234 [Jatropha curcas] |
| 3 |
Hb_000096_080 |
0.1748907089 |
- |
- |
PREDICTED: putative glycosyltransferase 7 [Jatropha curcas] |
| 4 |
Hb_000069_710 |
0.1881179224 |
- |
- |
PREDICTED: uncharacterized protein LOC105642890 [Jatropha curcas] |
| 5 |
Hb_000207_100 |
0.1974801251 |
- |
- |
PREDICTED: ABC transporter G family member 25 [Jatropha curcas] |
| 6 |
Hb_022174_010 |
0.2019240492 |
- |
- |
elicitor-responsive protein [Hevea brasiliensis] |
| 7 |
Hb_000943_030 |
0.2175810036 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_007810_100 |
0.2200487108 |
- |
- |
Synaptic glycoprotein SC2, putative [Ricinus communis] |
| 9 |
Hb_000684_130 |
0.2209066865 |
- |
- |
- |
| 10 |
Hb_002078_160 |
0.221322779 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 11 |
Hb_000056_040 |
0.2294578328 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31 [Jatropha curcas] |
| 12 |
Hb_000397_100 |
0.2331343429 |
- |
- |
hypothetical protein 31 [Hevea brasiliensis] |
| 13 |
Hb_000300_690 |
0.2364063605 |
- |
- |
hypothetical protein JCGZ_02719 [Jatropha curcas] |
| 14 |
Hb_007904_210 |
0.2385696798 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 1D [Jatropha curcas] |
| 15 |
Hb_003376_050 |
0.2408172863 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 16 |
Hb_007064_040 |
0.2410431494 |
- |
- |
hypothetical protein RCOM_0254640 [Ricinus communis] |
| 17 |
Hb_004375_130 |
0.2418885756 |
- |
- |
kinesin heavy chain, putative [Ricinus communis] |
| 18 |
Hb_003872_010 |
0.2433835006 |
- |
- |
hypothetical protein PRUPE_ppa013215mg [Prunus persica] |
| 19 |
Hb_000556_150 |
0.2450553218 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 10 [Populus euphratica] |
| 20 |
Hb_000933_060 |
0.2463364497 |
- |
- |
conserved hypothetical protein [Ricinus communis] |