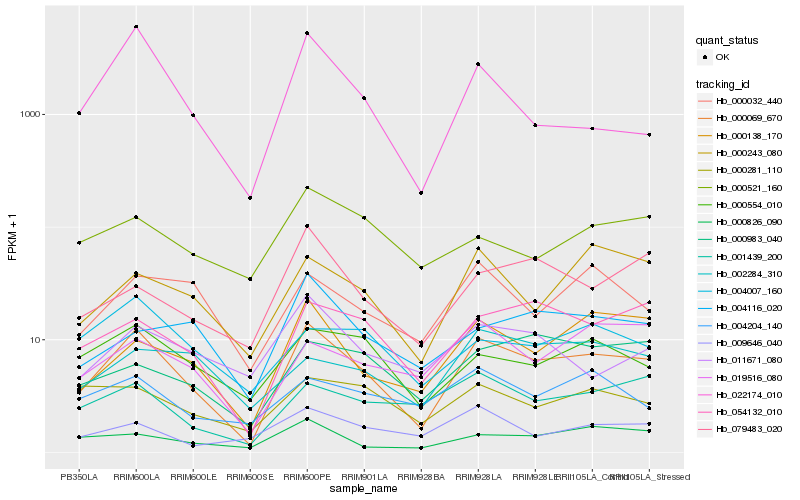
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000069_670 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642887 [Jatropha curcas] |
| 2 |
Hb_054132_010 |
0.1756704809 |
- |
- |
PREDICTED: transmembrane protein 136-like isoform X2 [Pyrus x bretschneideri] |
| 3 |
Hb_000138_170 |
0.176727647 |
- |
- |
putative plastid-lipid-associated protein, partial [Hevea brasiliensis] |
| 4 |
Hb_000243_080 |
0.1896647324 |
- |
- |
PREDICTED: beta-galactosidase 3 [Jatropha curcas] |
| 5 |
Hb_000554_010 |
0.1926600376 |
- |
- |
transporter, putative [Ricinus communis] |
| 6 |
Hb_001439_200 |
0.1980069103 |
- |
- |
Transmembrane protein Tmp21 precursor, putative [Ricinus communis] |
| 7 |
Hb_000826_090 |
0.1983869015 |
- |
- |
PREDICTED: cationic peroxidase 1-like [Populus euphratica] |
| 8 |
Hb_000032_440 |
0.2015755191 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_019516_080 |
0.203012561 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_079483_020 |
0.204374718 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 2-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_011671_080 |
0.2064319925 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase TDR [Jatropha curcas] |
| 12 |
Hb_000983_040 |
0.2072590337 |
- |
- |
PREDICTED: transmembrane protein 136 [Jatropha curcas] |
| 13 |
Hb_004116_020 |
0.208498752 |
- |
- |
fad NAD binding oxidoreductases, putative [Ricinus communis] |
| 14 |
Hb_004007_160 |
0.2096280161 |
- |
- |
PREDICTED: formin-like protein 6 [Jatropha curcas] |
| 15 |
Hb_022174_010 |
0.2099345147 |
- |
- |
elicitor-responsive protein [Hevea brasiliensis] |
| 16 |
Hb_004204_140 |
0.2125588897 |
transcription factor |
TF Family: C2H2 |
hypothetical protein RCOM_1470470 [Ricinus communis] |
| 17 |
Hb_000281_110 |
0.2134082079 |
- |
- |
PREDICTED: pumilio homolog 12 [Jatropha curcas] |
| 18 |
Hb_009646_040 |
0.2138682422 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_002284_310 |
0.214953203 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000521_160 |
0.2154184763 |
- |
- |
conserved hypothetical protein [Ricinus communis] |