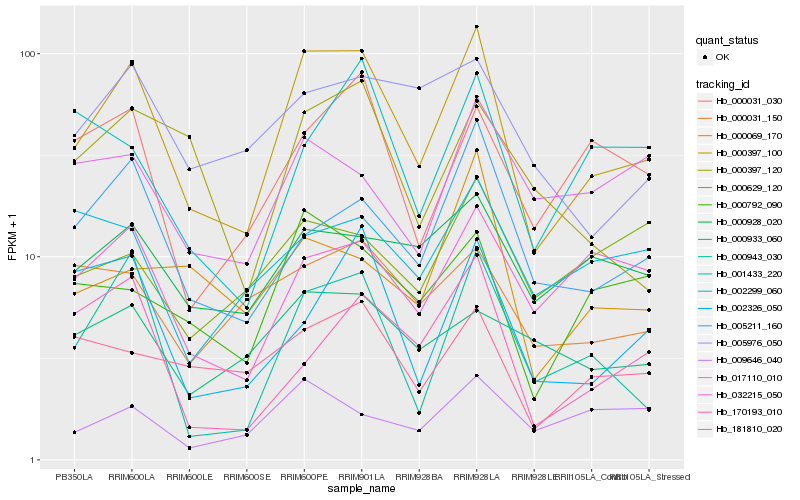
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000397_100 |
0.0 |
- |
- |
hypothetical protein 31 [Hevea brasiliensis] |
| 2 |
Hb_000943_030 |
0.1646852357 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_005211_160 |
0.1713349538 |
- |
- |
Protein PNS1, putative [Ricinus communis] |
| 4 |
Hb_000792_090 |
0.1793402986 |
- |
- |
3-beta-hydroxy-delta5-steroid dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_032215_050 |
0.1859522793 |
- |
- |
Xyloglucan galactosyltransferase KATAMARI1, putative [Ricinus communis] |
| 6 |
Hb_000928_020 |
0.1887977759 |
- |
- |
hypothetical protein CICLE_v10013682mg [Citrus clementina] |
| 7 |
Hb_000031_030 |
0.1894347778 |
- |
- |
PREDICTED: plasma membrane ATPase 4 [Jatropha curcas] |
| 8 |
Hb_181810_020 |
0.1898127838 |
- |
- |
- |
| 9 |
Hb_002326_050 |
0.1994952818 |
- |
- |
PREDICTED: uncharacterized protein LOC105633701 [Jatropha curcas] |
| 10 |
Hb_000031_150 |
0.2002051278 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 11 |
Hb_005976_050 |
0.2017530896 |
- |
- |
PREDICTED: protein RALF-like 34 [Jatropha curcas] |
| 12 |
Hb_009646_040 |
0.202120447 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_170193_010 |
0.2027491865 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 14 |
Hb_000933_060 |
0.2059625652 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000397_120 |
0.2065357664 |
- |
- |
PREDICTED: uncharacterized protein LOC105641234 [Jatropha curcas] |
| 16 |
Hb_001433_220 |
0.2093688907 |
- |
- |
protein phosphatase, putative [Ricinus communis] |
| 17 |
Hb_002299_060 |
0.210286952 |
transcription factor |
TF Family: FAR1 |
PREDICTED: probable receptor-like protein kinase At2g39360 [Jatropha curcas] |
| 18 |
Hb_000069_170 |
0.2104519712 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 18 [Jatropha curcas] |
| 19 |
Hb_017110_010 |
0.210864577 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At1g55760 [Jatropha curcas] |
| 20 |
Hb_000629_120 |
0.2126264279 |
- |
- |
Polynucleotidyl transferase, ribonuclease H-like superfamily protein [Theobroma cacao] |