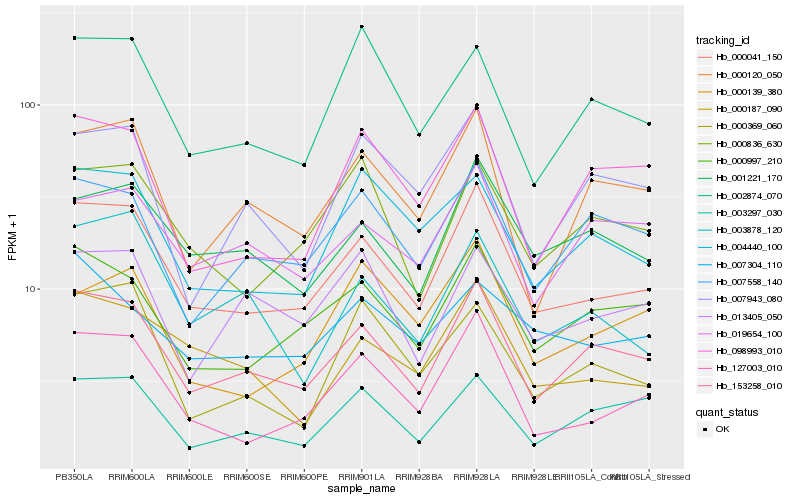
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000041_150 |
0.0 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |
| 2 |
Hb_153258_010 |
0.0763809387 |
- |
- |
PREDICTED: uncharacterized protein LOC105642325 [Jatropha curcas] |
| 3 |
Hb_127003_010 |
0.0864556591 |
- |
- |
PREDICTED: uncharacterized protein LOC105633613 [Jatropha curcas] |
| 4 |
Hb_000997_210 |
0.1051616682 |
- |
- |
PREDICTED: protein transport protein sec23-1 [Jatropha curcas] |
| 5 |
Hb_000187_090 |
0.1100794543 |
- |
- |
PREDICTED: magnesium transporter MRS2-3 [Jatropha curcas] |
| 6 |
Hb_001221_170 |
0.1132976926 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g50390, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000120_050 |
0.1199682126 |
- |
- |
PREDICTED: protein ELF4-LIKE 1 [Jatropha curcas] |
| 8 |
Hb_000836_630 |
0.123420871 |
- |
- |
PREDICTED: WD repeat-containing protein 44 [Jatropha curcas] |
| 9 |
Hb_004440_100 |
0.1245496035 |
- |
- |
PREDICTED: transmembrane protein 53 [Jatropha curcas] |
| 10 |
Hb_007558_140 |
0.1254686937 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 1-like [Jatropha curcas] |
| 11 |
Hb_098993_010 |
0.1296547057 |
- |
- |
PREDICTED: chaperone protein dnaJ 72 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002874_070 |
0.1327230363 |
- |
- |
PREDICTED: RNA-binding KH domain-containing protein PEPPER-like [Jatropha curcas] |
| 13 |
Hb_003878_120 |
0.133147251 |
transcription factor |
TF Family: MBF1 |
PREDICTED: multiprotein-bridging factor 1b-like [Elaeis guineensis] |
| 14 |
Hb_003297_030 |
0.1339535625 |
- |
- |
PREDICTED: uncharacterized protein LOC105640530 [Jatropha curcas] |
| 15 |
Hb_019654_100 |
0.1363364163 |
- |
- |
PREDICTED: tRNA pseudouridine synthase A, mitochondrial-like [Jatropha curcas] |
| 16 |
Hb_000369_060 |
0.1372924496 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 17 |
Hb_000139_380 |
0.1385723018 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_007943_080 |
0.1388264541 |
- |
- |
PREDICTED: cyclin-dependent kinase F-1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_013405_050 |
0.1393925439 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 [Jatropha curcas] |
| 20 |
Hb_007304_110 |
0.1395249761 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase 1 isoform X3 [Jatropha curcas] |