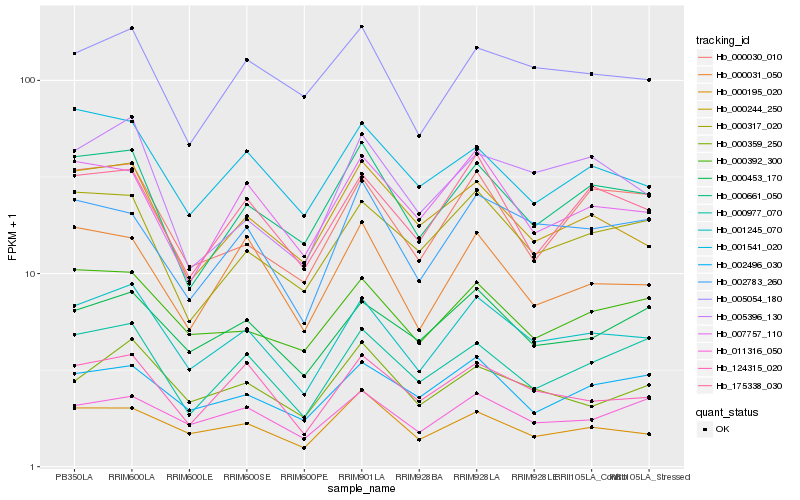
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001245_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633557 [Jatropha curcas] |
| 2 |
Hb_124315_020 |
0.0808043421 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_000661_050 |
0.0826000048 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 13C-like [Jatropha curcas] |
| 4 |
Hb_175338_030 |
0.0842575013 |
- |
- |
PREDICTED: uncharacterized protein LOC105648015 [Jatropha curcas] |
| 5 |
Hb_000453_170 |
0.0847861278 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_000392_300 |
0.0862744553 |
- |
- |
PREDICTED: RINT1-like protein MAG2 [Jatropha curcas] |
| 7 |
Hb_000244_250 |
0.0865987918 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 38 isoform X1 [Jatropha curcas] |
| 8 |
Hb_011316_050 |
0.0900112501 |
- |
- |
hypothetical protein CICLE_v10004643mg [Citrus clementina] |
| 9 |
Hb_002783_260 |
0.0903318672 |
- |
- |
hypothetical protein JCGZ_08219 [Jatropha curcas] |
| 10 |
Hb_000195_020 |
0.0905120271 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g19191, mitochondrial [Jatropha curcas] |
| 11 |
Hb_000317_020 |
0.0909166866 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex non-core subunit NAF1 [Jatropha curcas] |
| 12 |
Hb_005054_180 |
0.0911455599 |
- |
- |
coronatine-insensitive 1 [Hevea brasiliensis] |
| 13 |
Hb_000359_250 |
0.0913985919 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g26790, mitochondrial [Vitis vinifera] |
| 14 |
Hb_007757_110 |
0.0915204299 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5 [Jatropha curcas] |
| 15 |
Hb_000977_070 |
0.0923383279 |
- |
- |
PREDICTED: type II inositol 1,4,5-trisphosphate 5-phosphatase FRA3 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000031_050 |
0.0926431907 |
- |
- |
PREDICTED: uncharacterized protein LOC105637478 [Jatropha curcas] |
| 17 |
Hb_002496_030 |
0.0947556874 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g69350, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000030_010 |
0.0948863962 |
- |
- |
PREDICTED: uncharacterized protein LOC105632627 [Jatropha curcas] |
| 19 |
Hb_001541_020 |
0.095307109 |
- |
- |
PREDICTED: uncharacterized protein At3g49055 [Jatropha curcas] |
| 20 |
Hb_005396_130 |
0.0971188085 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |