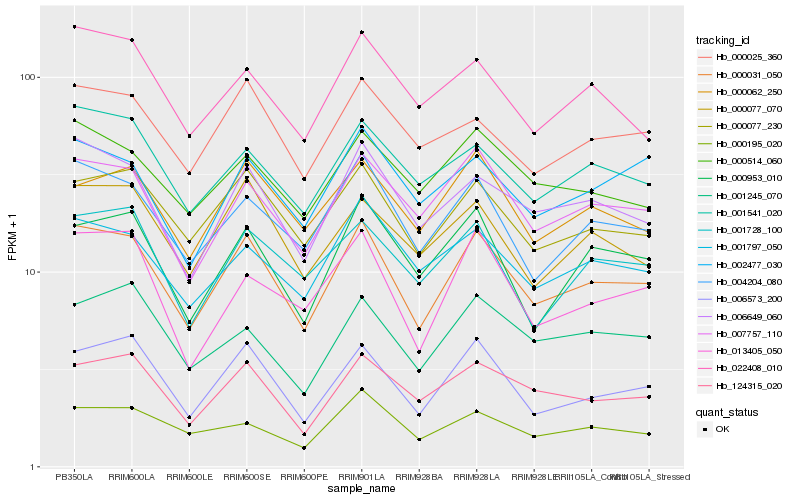
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000031_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637478 [Jatropha curcas] |
| 2 |
Hb_006573_200 |
0.0689301018 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g52850, chloroplastic [Jatropha curcas] |
| 3 |
Hb_006649_060 |
0.0718470303 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000077_230 |
0.0726696669 |
- |
- |
PREDICTED: uncharacterized protein LOC105646947 [Jatropha curcas] |
| 5 |
Hb_004204_080 |
0.0727584942 |
- |
- |
diacylglycerol kinase, theta, putative [Ricinus communis] |
| 6 |
Hb_007757_110 |
0.0753664789 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5 [Jatropha curcas] |
| 7 |
Hb_124315_020 |
0.0816139959 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_000953_010 |
0.0878078965 |
- |
- |
hypothetical protein CISIN_1g0425012mg, partial [Citrus sinensis] |
| 9 |
Hb_000077_070 |
0.088296572 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000025_360 |
0.0883149465 |
- |
- |
PREDICTED: protein SMG7 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001541_020 |
0.0889788351 |
- |
- |
PREDICTED: uncharacterized protein At3g49055 [Jatropha curcas] |
| 12 |
Hb_001797_050 |
0.0897017445 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 3-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000062_250 |
0.0907292803 |
- |
- |
PREDICTED: F-box protein SKIP14 [Jatropha curcas] |
| 14 |
Hb_013405_050 |
0.0918734805 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 [Jatropha curcas] |
| 15 |
Hb_000514_060 |
0.0923808818 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG isoform X1 [Jatropha curcas] |
| 16 |
Hb_001245_070 |
0.0926431907 |
- |
- |
PREDICTED: uncharacterized protein LOC105633557 [Jatropha curcas] |
| 17 |
Hb_000195_020 |
0.0935646131 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g19191, mitochondrial [Jatropha curcas] |
| 18 |
Hb_001728_100 |
0.0937995546 |
- |
- |
hypothetical protein JCGZ_03429 [Jatropha curcas] |
| 19 |
Hb_022408_010 |
0.0948563624 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3 [Jatropha curcas] |
| 20 |
Hb_002477_030 |
0.0967880119 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105631404 isoform X1 [Jatropha curcas] |