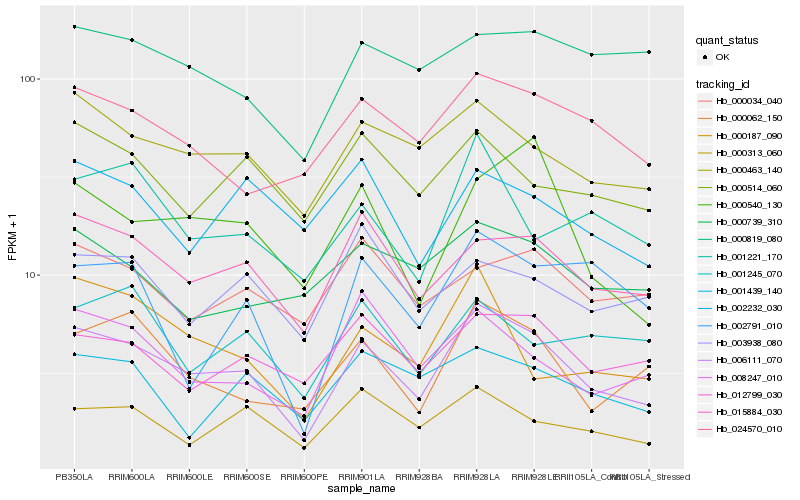
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006111_070 |
0.0 |
- |
- |
hypothetical protein POPTR_0008s07660g [Populus trichocarpa] |
| 2 |
Hb_015884_030 |
0.127666186 |
- |
- |
PREDICTED: uncharacterized protein LOC105647778 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000463_140 |
0.1302190798 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |
| 4 |
Hb_000062_150 |
0.1356291625 |
- |
- |
PREDICTED: probable carboxylesterase 16 [Jatropha curcas] |
| 5 |
Hb_024570_010 |
0.1414941838 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 6 |
Hb_012799_030 |
0.1464648972 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g55630-like [Jatropha curcas] |
| 7 |
Hb_000313_060 |
0.1473776306 |
- |
- |
hypothetical protein [Beta vulgaris subsp. vulgaris] |
| 8 |
Hb_001221_170 |
0.1493340947 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g50390, chloroplastic [Jatropha curcas] |
| 9 |
Hb_008247_010 |
0.1509525298 |
- |
- |
PREDICTED: leucine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 10 |
Hb_000540_130 |
0.1527380583 |
transcription factor |
TF Family: Pseudo ARR-B |
sensory transduction histidine kinase, putative [Ricinus communis] |
| 11 |
Hb_000514_060 |
0.1527561577 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG isoform X1 [Jatropha curcas] |
| 12 |
Hb_000187_090 |
0.1560079462 |
- |
- |
PREDICTED: magnesium transporter MRS2-3 [Jatropha curcas] |
| 13 |
Hb_001439_140 |
0.1618781357 |
- |
- |
pattern formation protein, putative [Ricinus communis] |
| 14 |
Hb_002232_030 |
0.1624019377 |
- |
- |
Pinin, putative [Ricinus communis] |
| 15 |
Hb_000739_310 |
0.1626241929 |
- |
- |
PREDICTED: uncharacterized protein LOC105643091 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001245_070 |
0.1626349841 |
- |
- |
PREDICTED: uncharacterized protein LOC105633557 [Jatropha curcas] |
| 17 |
Hb_000034_040 |
0.1644847122 |
- |
- |
PREDICTED: histidine--tRNA ligase [Jatropha curcas] |
| 18 |
Hb_000819_080 |
0.1662632332 |
- |
- |
Plastid-lipid-associated protein, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_002791_010 |
0.1662837048 |
- |
- |
PREDICTED: general transcription factor 3C polypeptide 3-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_003938_080 |
0.1681012772 |
- |
- |
PREDICTED: uncharacterized protein LOC105644554 [Jatropha curcas] |