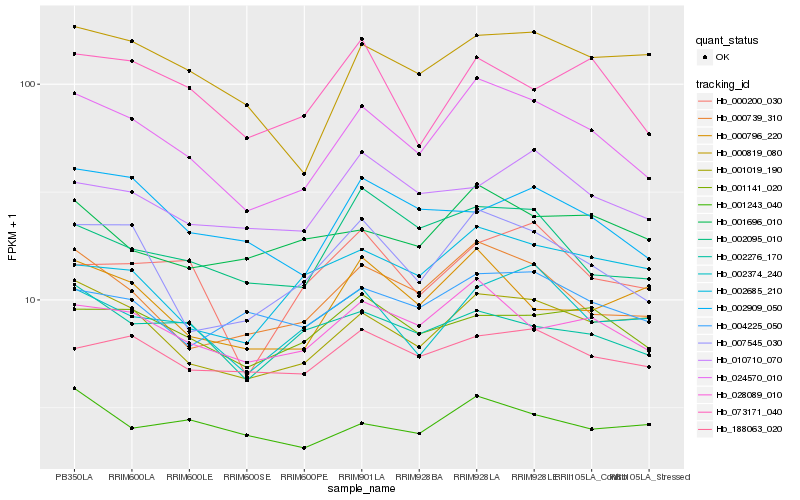
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_024570_010 |
0.0 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 2 |
Hb_000739_310 |
0.0786179975 |
- |
- |
PREDICTED: uncharacterized protein LOC105643091 isoform X1 [Jatropha curcas] |
| 3 |
Hb_007545_030 |
0.0802455902 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_028089_010 |
0.0832375201 |
- |
- |
PREDICTED: homoserine dehydrogenase [Jatropha curcas] |
| 5 |
Hb_001019_190 |
0.0849969012 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002374_240 |
0.1009778922 |
- |
- |
nucellin, putative [Ricinus communis] |
| 7 |
Hb_000819_080 |
0.1045487053 |
- |
- |
Plastid-lipid-associated protein, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_002095_010 |
0.105952314 |
- |
- |
PREDICTED: glutamate receptor 3.7 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001141_020 |
0.1067771865 |
- |
- |
PREDICTED: stress-induced-phosphoprotein 1 [Jatropha curcas] |
| 10 |
Hb_000796_220 |
0.1081622319 |
- |
- |
PREDICTED: uncharacterized protein LOC105642644 isoform X3 [Jatropha curcas] |
| 11 |
Hb_073171_040 |
0.1088122276 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 12 |
Hb_004225_050 |
0.10989153 |
transcription factor |
TF Family: MYB-related |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_188063_020 |
0.1114691166 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002909_050 |
0.1118747133 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002276_170 |
0.1118871087 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002685_210 |
0.1126666632 |
- |
- |
PREDICTED: BTB/POZ and MATH domain-containing protein 4 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001696_010 |
0.1130540725 |
- |
- |
PREDICTED: serine/threonine-protein kinase 38-like isoform X3 [Jatropha curcas] |
| 18 |
Hb_000200_030 |
0.113257056 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATC, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001243_040 |
0.1133057528 |
- |
- |
PREDICTED: uncharacterized protein LOC104104316 [Nicotiana tomentosiformis] |
| 20 |
Hb_010710_070 |
0.1135863057 |
- |
- |
PREDICTED: protein TOC75-3, chloroplastic-like [Populus euphratica] |