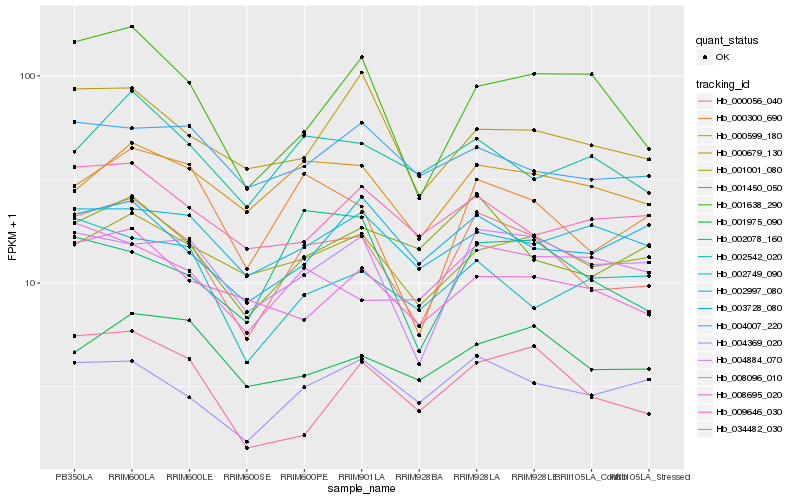
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000056_040 |
0.0 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31 [Jatropha curcas] |
| 2 |
Hb_000300_690 |
0.1013643983 |
- |
- |
hypothetical protein JCGZ_02719 [Jatropha curcas] |
| 3 |
Hb_004369_020 |
0.1122702309 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g63330-like [Jatropha curcas] |
| 4 |
Hb_004884_070 |
0.1131918774 |
- |
- |
PREDICTED: ethylene-overproduction protein 1 [Jatropha curcas] |
| 5 |
Hb_001450_050 |
0.1172204258 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000599_180 |
0.1233900808 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 7 |
Hb_001638_290 |
0.1240934296 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_001001_080 |
0.124812564 |
- |
- |
PREDICTED: uncharacterized protein LOC105633661 [Jatropha curcas] |
| 9 |
Hb_008695_020 |
0.1295302041 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 1 [Jatropha curcas] |
| 10 |
Hb_002542_020 |
0.130291458 |
- |
- |
PREDICTED: uncharacterized protein LOC105634003 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000679_130 |
0.1321932976 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 12 |
Hb_001975_090 |
0.1330749255 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 13 |
Hb_009646_030 |
0.1341674713 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003728_080 |
0.1353506575 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3-like [Jatropha curcas] |
| 15 |
Hb_002749_090 |
0.1360937888 |
- |
- |
- |
| 16 |
Hb_004007_220 |
0.1364787604 |
- |
- |
PREDICTED: casein kinase II subunit alpha [Jatropha curcas] |
| 17 |
Hb_008096_010 |
0.1365481862 |
- |
- |
hypothetical protein MTR_1g045660 [Medicago truncatula] |
| 18 |
Hb_034482_030 |
0.13793463 |
- |
- |
PREDICTED: histidine--tRNA ligase [Jatropha curcas] |
| 19 |
Hb_002997_080 |
0.1389767941 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |
| 20 |
Hb_002078_160 |
0.1399098296 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |