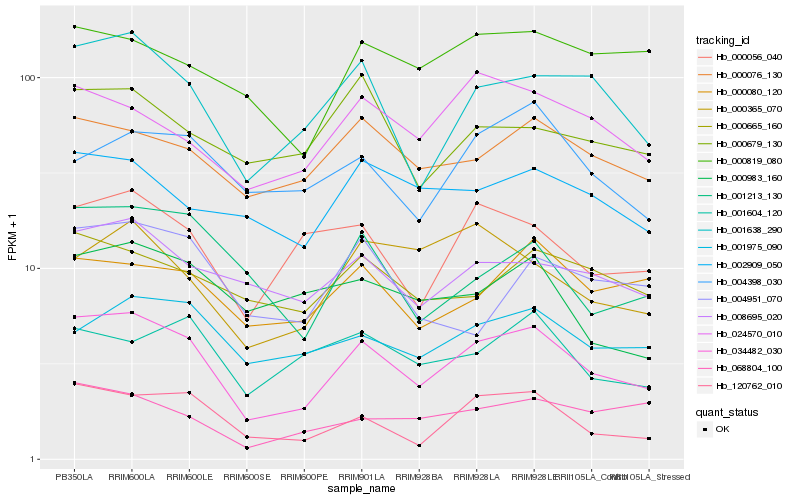
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_034482_030 |
0.0 |
- |
- |
PREDICTED: histidine--tRNA ligase [Jatropha curcas] |
| 2 |
Hb_001638_290 |
0.1135253737 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_120762_010 |
0.116409578 |
- |
- |
- |
| 4 |
Hb_000056_040 |
0.13793463 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31 [Jatropha curcas] |
| 5 |
Hb_008695_020 |
0.1384627968 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 1 [Jatropha curcas] |
| 6 |
Hb_000076_130 |
0.1411619289 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |
| 7 |
Hb_001213_130 |
0.1432378651 |
- |
- |
PREDICTED: fructokinase-like 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_024570_010 |
0.1458254563 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 9 |
Hb_001975_090 |
0.1466239071 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 10 |
Hb_000080_120 |
0.146825099 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62350-like [Jatropha curcas] |
| 11 |
Hb_068804_100 |
0.1484858363 |
- |
- |
PREDICTED: surfeit locus protein 6 [Jatropha curcas] |
| 12 |
Hb_000665_160 |
0.1496780152 |
- |
- |
PREDICTED: cysteine--tRNA ligase, cytoplasmic isoform X1 [Jatropha curcas] |
| 13 |
Hb_002909_050 |
0.1508000389 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001604_120 |
0.1537982551 |
- |
- |
PREDICTED: hemK methyltransferase family member 1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_004398_030 |
0.1555528685 |
- |
- |
PREDICTED: chloroplast processing peptidase [Jatropha curcas] |
| 16 |
Hb_000365_070 |
0.1556802824 |
- |
- |
PREDICTED: SWR1 complex subunit 2 [Jatropha curcas] |
| 17 |
Hb_004951_070 |
0.1561897387 |
desease resistance |
Gene Name: AAA |
Stage III sporulation protein AA, putative [Ricinus communis] |
| 18 |
Hb_000983_160 |
0.1562564646 |
- |
- |
hypothetical protein B456_009G199800 [Gossypium raimondii] |
| 19 |
Hb_000679_130 |
0.156351095 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 20 |
Hb_000819_080 |
0.156611302 |
- |
- |
Plastid-lipid-associated protein, chloroplast precursor, putative [Ricinus communis] |