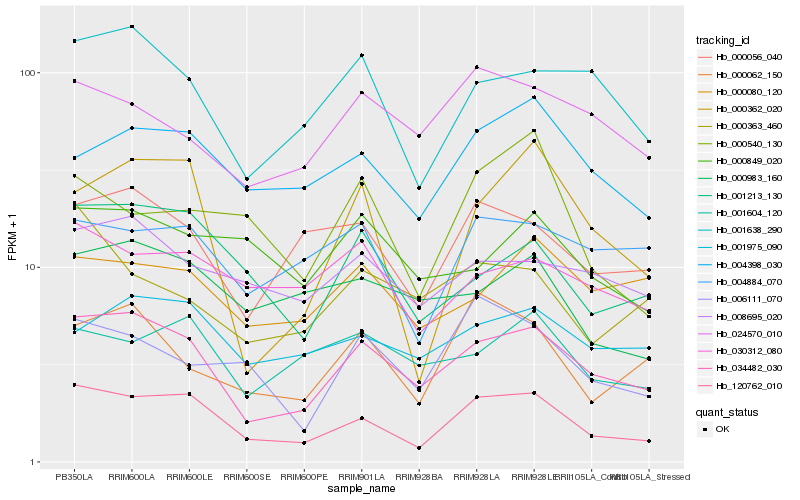
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_120762_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_034482_030 |
0.116409578 |
- |
- |
PREDICTED: histidine--tRNA ligase [Jatropha curcas] |
| 3 |
Hb_001213_130 |
0.1503656368 |
- |
- |
PREDICTED: fructokinase-like 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_004398_030 |
0.1596928916 |
- |
- |
PREDICTED: chloroplast processing peptidase [Jatropha curcas] |
| 5 |
Hb_000056_040 |
0.1647890736 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31 [Jatropha curcas] |
| 6 |
Hb_000362_020 |
0.1676848213 |
transcription factor |
TF Family: TCP |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_006111_070 |
0.1718787547 |
- |
- |
hypothetical protein POPTR_0008s07660g [Populus trichocarpa] |
| 8 |
Hb_001638_290 |
0.1724463164 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |
| 9 |
Hb_030312_080 |
0.1735818009 |
- |
- |
calcium-dependent protein kinase 6 [Hevea brasiliensis] |
| 10 |
Hb_000540_130 |
0.1756143261 |
transcription factor |
TF Family: Pseudo ARR-B |
sensory transduction histidine kinase, putative [Ricinus communis] |
| 11 |
Hb_004884_070 |
0.1761936214 |
- |
- |
PREDICTED: ethylene-overproduction protein 1 [Jatropha curcas] |
| 12 |
Hb_000062_150 |
0.1768163986 |
- |
- |
PREDICTED: probable carboxylesterase 16 [Jatropha curcas] |
| 13 |
Hb_000983_160 |
0.1776201215 |
- |
- |
hypothetical protein B456_009G199800 [Gossypium raimondii] |
| 14 |
Hb_001604_120 |
0.1782746125 |
- |
- |
PREDICTED: hemK methyltransferase family member 1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001975_090 |
0.1805857068 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 16 |
Hb_008695_020 |
0.1839919199 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 1 [Jatropha curcas] |
| 17 |
Hb_000080_120 |
0.1923276855 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62350-like [Jatropha curcas] |
| 18 |
Hb_000849_020 |
0.1934446326 |
- |
- |
PREDICTED: uncharacterized protein LOC105644228 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000363_460 |
0.1937144093 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial isoform X1 [Jatropha curcas] |
| 20 |
Hb_024570_010 |
0.194768255 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |