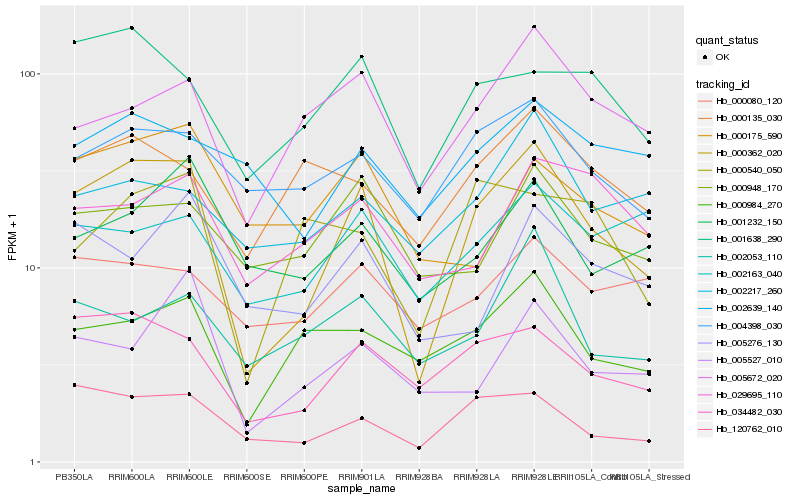
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000362_020 |
0.0 |
transcription factor |
TF Family: TCP |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_034482_030 |
0.163580886 |
- |
- |
PREDICTED: histidine--tRNA ligase [Jatropha curcas] |
| 3 |
Hb_120762_010 |
0.1676848213 |
- |
- |
- |
| 4 |
Hb_001638_290 |
0.1911636303 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_005672_020 |
0.1963457856 |
- |
- |
PREDICTED: 50S ribosomal protein L6, chloroplastic [Jatropha curcas] |
| 6 |
Hb_004398_030 |
0.1994093643 |
- |
- |
PREDICTED: chloroplast processing peptidase [Jatropha curcas] |
| 7 |
Hb_000984_270 |
0.1995113142 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001232_150 |
0.2071033643 |
- |
- |
PREDICTED: MAR-binding filament-like protein 1-1 [Jatropha curcas] |
| 9 |
Hb_005527_010 |
0.2074107765 |
- |
- |
PREDICTED: cell division protein FtsY homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_005276_130 |
0.2075160004 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000948_170 |
0.2115170466 |
- |
- |
hypothetical protein JCGZ_06749 [Jatropha curcas] |
| 12 |
Hb_000540_050 |
0.213394289 |
- |
- |
PREDICTED: 125 kDa kinesin-related protein-like [Jatropha curcas] |
| 13 |
Hb_000175_590 |
0.2136422592 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000135_030 |
0.2141552734 |
- |
- |
PREDICTED: uncharacterized protein LOC105634729 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002163_040 |
0.2142147306 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002053_110 |
0.2146594542 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_000080_120 |
0.2168246444 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62350-like [Jatropha curcas] |
| 18 |
Hb_029695_110 |
0.2179633401 |
- |
- |
hypothetical protein CICLE_v10021605mg [Citrus clementina] |
| 19 |
Hb_002217_260 |
0.2180070899 |
- |
- |
PREDICTED: uncharacterized protein LOC105643577 [Jatropha curcas] |
| 20 |
Hb_002639_140 |
0.2185304955 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase-like 5 [Jatropha curcas] |