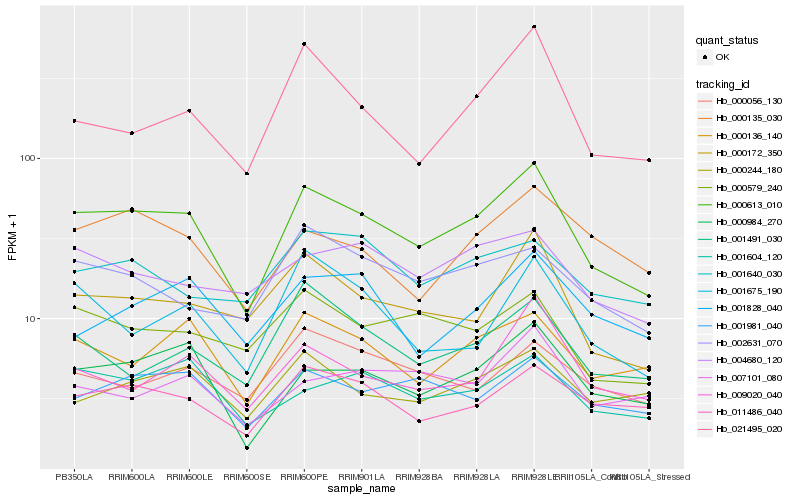
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000613_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647991 [Jatropha curcas] |
| 2 |
Hb_000984_270 |
0.106268066 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000172_350 |
0.1191977306 |
- |
- |
PREDICTED: uncharacterized protein LOC105650963 [Jatropha curcas] |
| 4 |
Hb_000136_140 |
0.1212353294 |
- |
- |
PREDICTED: uncharacterized protein LOC105648960 [Jatropha curcas] |
| 5 |
Hb_011486_040 |
0.1226074225 |
- |
- |
PREDICTED: protein trichome birefringence-like 13 [Jatropha curcas] |
| 6 |
Hb_001981_040 |
0.130410389 |
transcription factor |
TF Family: PHD |
PREDICTED: origin of replication complex subunit 1B-like [Jatropha curcas] |
| 7 |
Hb_009020_040 |
0.1315754647 |
- |
- |
PREDICTED: triacylglycerol lipase 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001604_120 |
0.1315902041 |
- |
- |
PREDICTED: hemK methyltransferase family member 1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000579_240 |
0.1331647873 |
- |
- |
phosphatidylinositol transporter, putative [Ricinus communis] |
| 10 |
Hb_021495_020 |
0.1339319084 |
- |
- |
chitinase, putative [Ricinus communis] |
| 11 |
Hb_000135_030 |
0.1339932151 |
- |
- |
PREDICTED: uncharacterized protein LOC105634729 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000244_180 |
0.1361215771 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002631_070 |
0.1370324109 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860-like [Jatropha curcas] |
| 14 |
Hb_001675_190 |
0.1390770551 |
- |
- |
PREDICTED: probable plastidic glucose transporter 3 [Jatropha curcas] |
| 15 |
Hb_001491_030 |
0.1396168178 |
- |
- |
PREDICTED: uncharacterized protein LOC105649804 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004680_120 |
0.1398869899 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 1 [Jatropha curcas] |
| 17 |
Hb_001828_040 |
0.1451046673 |
- |
- |
PREDICTED: F-box protein SKIP14 [Jatropha curcas] |
| 18 |
Hb_000056_130 |
0.1465791465 |
- |
- |
Sucrose Transporter 2C [Hevea brasiliensis subsp. brasiliensis] |
| 19 |
Hb_007101_080 |
0.1466422355 |
transcription factor |
TF Family: GNAT |
GCN5-related N-acetyltransferase family protein [Populus trichocarpa] |
| 20 |
Hb_001640_030 |
0.1467869797 |
- |
- |
PREDICTED: uncharacterized protein LOC105630021 [Jatropha curcas] |