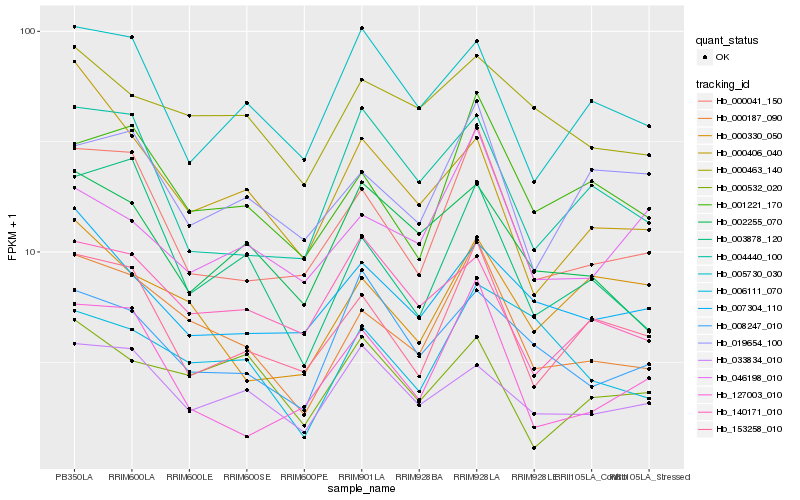
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000187_090 |
0.0 |
- |
- |
PREDICTED: magnesium transporter MRS2-3 [Jatropha curcas] |
| 2 |
Hb_000041_150 |
0.1100794543 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |
| 3 |
Hb_001221_170 |
0.1226260783 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g50390, chloroplastic [Jatropha curcas] |
| 4 |
Hb_003878_120 |
0.1226828946 |
transcription factor |
TF Family: MBF1 |
PREDICTED: multiprotein-bridging factor 1b-like [Elaeis guineensis] |
| 5 |
Hb_153258_010 |
0.1334313309 |
- |
- |
PREDICTED: uncharacterized protein LOC105642325 [Jatropha curcas] |
| 6 |
Hb_000463_140 |
0.1514939399 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |
| 7 |
Hb_006111_070 |
0.1560079462 |
- |
- |
hypothetical protein POPTR_0008s07660g [Populus trichocarpa] |
| 8 |
Hb_002255_070 |
0.1575059369 |
- |
- |
PREDICTED: isoleucine--tRNA ligase, cytoplasmic-like [Camelina sativa] |
| 9 |
Hb_033834_010 |
0.1579694574 |
- |
- |
PREDICTED: leucine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 10 |
Hb_007304_110 |
0.1595589055 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase 1 isoform X3 [Jatropha curcas] |
| 11 |
Hb_019654_100 |
0.1610475015 |
- |
- |
PREDICTED: tRNA pseudouridine synthase A, mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_000406_040 |
0.1612703353 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: FRIGIDA-like protein 5 [Jatropha curcas] |
| 13 |
Hb_127003_010 |
0.1625496352 |
- |
- |
PREDICTED: uncharacterized protein LOC105633613 [Jatropha curcas] |
| 14 |
Hb_140171_010 |
0.1641496916 |
- |
- |
PREDICTED: magnesium transporter MRS2-3 [Jatropha curcas] |
| 15 |
Hb_000532_020 |
0.1659480434 |
- |
- |
beta-tubulin [Bouteloua eriopoda] |
| 16 |
Hb_000330_050 |
0.1659569968 |
- |
- |
Protein yrdA, putative [Ricinus communis] |
| 17 |
Hb_046198_010 |
0.1661229895 |
- |
- |
PREDICTED: NADPH-dependent pterin aldehyde reductase [Jatropha curcas] |
| 18 |
Hb_004440_100 |
0.1662570572 |
- |
- |
PREDICTED: transmembrane protein 53 [Jatropha curcas] |
| 19 |
Hb_005730_030 |
0.1673195148 |
- |
- |
eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 20 |
Hb_008247_010 |
0.1694752706 |
- |
- |
PREDICTED: leucine--tRNA ligase, cytoplasmic [Jatropha curcas] |