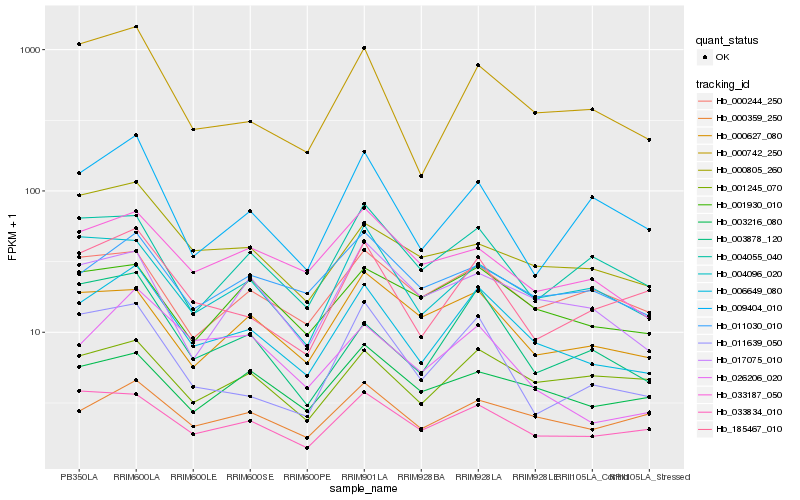
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006649_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640496 [Jatropha curcas] |
| 2 |
Hb_000742_250 |
0.108970581 |
- |
- |
Tubulin beta-6 chain [Gossypium arboreum] |
| 3 |
Hb_185467_010 |
0.1222389886 |
- |
- |
- |
| 4 |
Hb_033834_010 |
0.1241223469 |
- |
- |
PREDICTED: leucine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 5 |
Hb_000359_250 |
0.1310186308 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g26790, mitochondrial [Vitis vinifera] |
| 6 |
Hb_003878_120 |
0.1316220825 |
transcription factor |
TF Family: MBF1 |
PREDICTED: multiprotein-bridging factor 1b-like [Elaeis guineensis] |
| 7 |
Hb_004096_020 |
0.1316231397 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like, partial [Fragaria vesca subsp. vesca] |
| 8 |
Hb_026206_020 |
0.133008656 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At2g04740 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001245_070 |
0.1370967383 |
- |
- |
PREDICTED: uncharacterized protein LOC105633557 [Jatropha curcas] |
| 10 |
Hb_033187_050 |
0.1375088667 |
- |
- |
PREDICTED: exocyst complex component SEC15B [Jatropha curcas] |
| 11 |
Hb_011030_010 |
0.1378348006 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |
| 12 |
Hb_011639_050 |
0.1393291776 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001930_010 |
0.1401493783 |
- |
- |
PREDICTED: uncharacterized protein LOC105633074 [Jatropha curcas] |
| 14 |
Hb_009404_010 |
0.140333868 |
- |
- |
PREDICTED: casein kinase II subunit alpha-2-like [Glycine max] |
| 15 |
Hb_000627_080 |
0.1409833623 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 16 |
Hb_003216_080 |
0.1416926692 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g65560-like [Jatropha curcas] |
| 17 |
Hb_017075_010 |
0.1418056954 |
transcription factor |
TF Family: SNF2 |
PREDICTED: putative SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3-like 1 [Jatropha curcas] |
| 18 |
Hb_000244_250 |
0.1436711727 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 38 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004055_040 |
0.1437412361 |
- |
- |
Phospholipid-transporting ATPase, putative [Ricinus communis] |
| 20 |
Hb_000805_260 |
0.1444758918 |
- |
- |
PREDICTED: uncharacterized protein LOC105650295 isoform X1 [Jatropha curcas] |