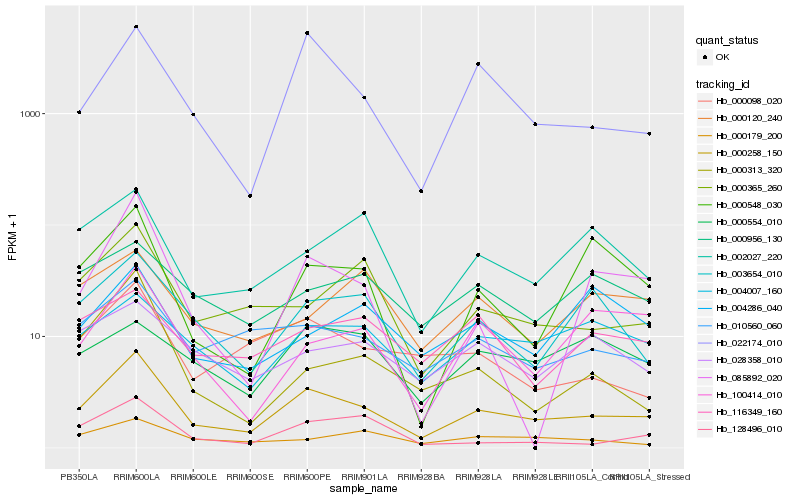
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000258_150 |
0.0 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 11-like [Jatropha curcas] |
| 2 |
Hb_010560_060 |
0.1789410362 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |
| 3 |
Hb_022174_010 |
0.181846249 |
- |
- |
elicitor-responsive protein [Hevea brasiliensis] |
| 4 |
Hb_003654_010 |
0.1891767951 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP650 [Jatropha curcas] |
| 5 |
Hb_000365_260 |
0.1929785349 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004007_160 |
0.1943417164 |
- |
- |
PREDICTED: formin-like protein 6 [Jatropha curcas] |
| 7 |
Hb_000098_020 |
0.1978833766 |
transcription factor |
TF Family: GRAS |
GRAS family transcription factor family protein [Theobroma cacao] |
| 8 |
Hb_128496_010 |
0.2034794801 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_002027_220 |
0.2047921469 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 10 |
Hb_100414_010 |
0.2083310001 |
rubber biosynthesis |
Gene Name: Farnesyl diphosphate synthase |
farnesyl pyrophosphate synthase [Hevea brasiliensis] |
| 11 |
Hb_000179_200 |
0.208478043 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_000548_030 |
0.2116881398 |
rubber biosynthesis |
Gene Name: CPT5 |
PREDICTED: dehydrodolichyl diphosphate synthase 6-like [Beta vulgaris subsp. vulgaris] |
| 13 |
Hb_028358_010 |
0.211785466 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000956_130 |
0.2151885408 |
- |
- |
hypothetical protein POPTR_0002s12620g [Populus trichocarpa] |
| 15 |
Hb_116349_160 |
0.2166249675 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 16 |
Hb_000313_320 |
0.217472286 |
- |
- |
PREDICTED: protein kinase PINOID [Jatropha curcas] |
| 17 |
Hb_085892_020 |
0.2219727112 |
- |
- |
- |
| 18 |
Hb_000554_010 |
0.2251502852 |
- |
- |
transporter, putative [Ricinus communis] |
| 19 |
Hb_000120_240 |
0.2279100122 |
- |
- |
PREDICTED: uncharacterized protein LOC105630166 [Jatropha curcas] |
| 20 |
Hb_004286_040 |
0.2279135804 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH74 isoform X1 [Jatropha curcas] |