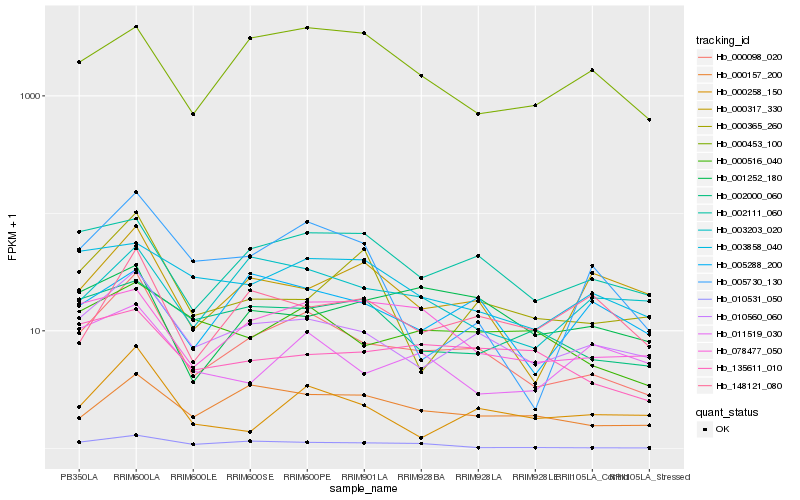
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000098_020 |
0.0 |
transcription factor |
TF Family: GRAS |
GRAS family transcription factor family protein [Theobroma cacao] |
| 2 |
Hb_010560_060 |
0.1284216646 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |
| 3 |
Hb_002111_060 |
0.1795043854 |
- |
- |
PREDICTED: toll-like receptor 13 [Jatropha curcas] |
| 4 |
Hb_000157_200 |
0.1895014418 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 isoform X3 [Eucalyptus grandis] |
| 5 |
Hb_000516_040 |
0.1895338138 |
- |
- |
PREDICTED: uncharacterized protein LOC105649502 isoform X2 [Jatropha curcas] |
| 6 |
Hb_078477_050 |
0.1966989107 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105648332 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000258_150 |
0.1978833766 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 11-like [Jatropha curcas] |
| 8 |
Hb_148121_080 |
0.2037178924 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SHOOT GRAVITROPISM 5 [Populus euphratica] |
| 9 |
Hb_003203_020 |
0.2039910296 |
- |
- |
unknown [Lotus japonicus] |
| 10 |
Hb_135611_010 |
0.2072685422 |
- |
- |
PREDICTED: uncharacterized protein LOC105142085 [Populus euphratica] |
| 11 |
Hb_003858_040 |
0.2113001399 |
- |
- |
hypothetical protein POPTR_0007s14590g [Populus trichocarpa] |
| 12 |
Hb_000453_100 |
0.212033795 |
- |
- |
PREDICTED: metallothionein-like protein 1 [Sesamum indicum] |
| 13 |
Hb_000317_330 |
0.2125125921 |
- |
- |
PREDICTED: RNA-binding protein 38-like [Jatropha curcas] |
| 14 |
Hb_005288_200 |
0.2129119219 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 15 |
Hb_001252_180 |
0.2142685336 |
desease resistance |
Gene Name: DUF3542 |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002000_060 |
0.2157801542 |
- |
- |
PREDICTED: protein LIKE COV 2 [Jatropha curcas] |
| 17 |
Hb_005730_130 |
0.2185879025 |
- |
- |
PRA1 family protein [Theobroma cacao] |
| 18 |
Hb_011519_030 |
0.2189643307 |
- |
- |
PREDICTED: uncharacterized protein At3g17950 [Jatropha curcas] |
| 19 |
Hb_010531_050 |
0.2193508043 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000365_260 |
0.2205498273 |
- |
- |
conserved hypothetical protein [Ricinus communis] |