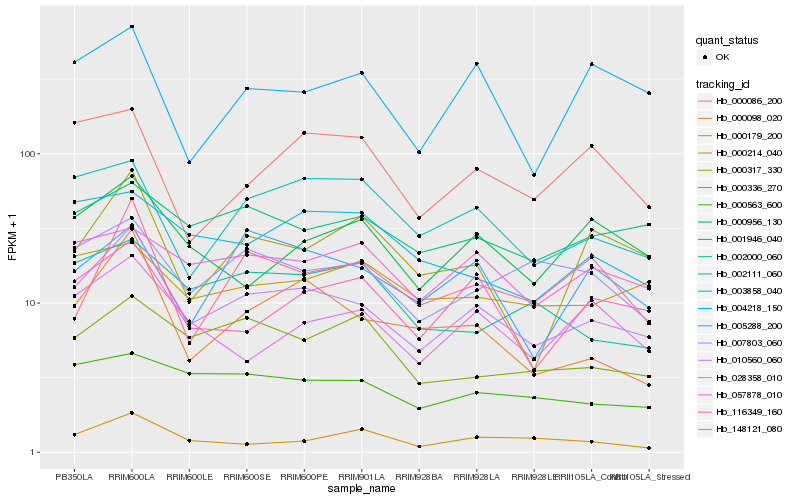
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010560_060 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |
| 2 |
Hb_000098_020 |
0.1284216646 |
transcription factor |
TF Family: GRAS |
GRAS family transcription factor family protein [Theobroma cacao] |
| 3 |
Hb_116349_160 |
0.148584501 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 4 |
Hb_028358_010 |
0.148979283 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_007803_060 |
0.1490772235 |
- |
- |
PREDICTED: uncharacterized protein LOC105632794 [Jatropha curcas] |
| 6 |
Hb_002111_060 |
0.1530272674 |
- |
- |
PREDICTED: toll-like receptor 13 [Jatropha curcas] |
| 7 |
Hb_000563_600 |
0.1544739722 |
- |
- |
PREDICTED: uncharacterized protein LOC104428079 [Eucalyptus grandis] |
| 8 |
Hb_000956_130 |
0.1564219053 |
- |
- |
hypothetical protein POPTR_0002s12620g [Populus trichocarpa] |
| 9 |
Hb_001946_040 |
0.1580063559 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_057878_010 |
0.1589340001 |
- |
- |
hypothetical protein VITISV_036304 [Vitis vinifera] |
| 11 |
Hb_000336_270 |
0.1589499696 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000317_330 |
0.163167418 |
- |
- |
PREDICTED: RNA-binding protein 38-like [Jatropha curcas] |
| 13 |
Hb_000179_200 |
0.1643234952 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_004218_150 |
0.1648711157 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 15 |
Hb_148121_080 |
0.1650296685 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SHOOT GRAVITROPISM 5 [Populus euphratica] |
| 16 |
Hb_005288_200 |
0.1663439545 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 17 |
Hb_002000_060 |
0.1665304875 |
- |
- |
PREDICTED: protein LIKE COV 2 [Jatropha curcas] |
| 18 |
Hb_003858_040 |
0.1667579143 |
- |
- |
hypothetical protein POPTR_0007s14590g [Populus trichocarpa] |
| 19 |
Hb_000214_040 |
0.1695825254 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000086_200 |
0.169874062 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |