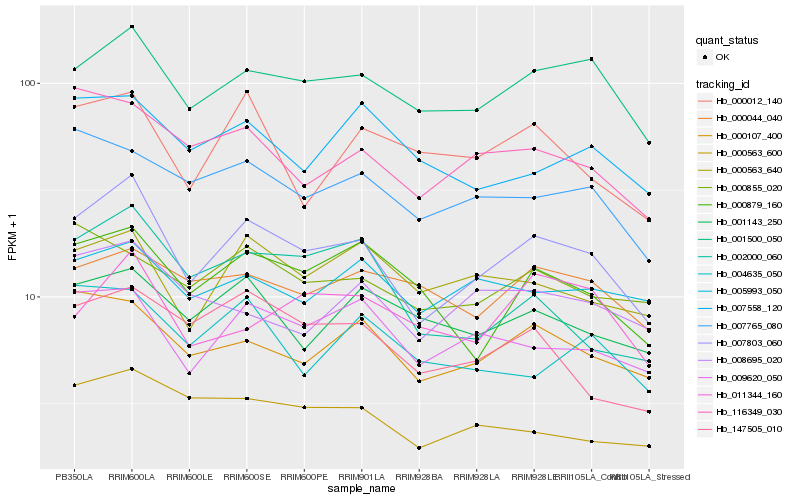
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007803_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632794 [Jatropha curcas] |
| 2 |
Hb_001500_050 |
0.0917128828 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 3 |
Hb_009620_050 |
0.1079453909 |
- |
- |
PREDICTED: protein unc-45 homolog A [Jatropha curcas] |
| 4 |
Hb_000107_400 |
0.1097610156 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46870-like [Jatropha curcas] |
| 5 |
Hb_116349_030 |
0.1161957089 |
- |
- |
PREDICTED: zinc finger MYND domain-containing protein 15 isoform X3 [Jatropha curcas] |
| 6 |
Hb_000879_160 |
0.1191156829 |
- |
- |
PREDICTED: F-box protein At3g12350 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000563_640 |
0.1203905032 |
- |
- |
PREDICTED: chaperone protein dnaJ 49 [Jatropha curcas] |
| 8 |
Hb_007765_080 |
0.1206971497 |
- |
- |
PREDICTED: uncharacterized protein LOC105631001 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000563_600 |
0.1210537904 |
- |
- |
PREDICTED: uncharacterized protein LOC104428079 [Eucalyptus grandis] |
| 10 |
Hb_147505_010 |
0.1232133555 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 11 |
Hb_001143_250 |
0.1263653998 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3C isoform X1 [Jatropha curcas] |
| 12 |
Hb_005993_050 |
0.1277845466 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 13 |
Hb_002000_060 |
0.1283933094 |
- |
- |
PREDICTED: protein LIKE COV 2 [Jatropha curcas] |
| 14 |
Hb_008695_020 |
0.1290846658 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 1 [Jatropha curcas] |
| 15 |
Hb_000012_140 |
0.1300186517 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_007558_120 |
0.1316333614 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 17 |
Hb_000855_020 |
0.1318532872 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X1 [Populus euphratica] |
| 18 |
Hb_011344_160 |
0.1336929223 |
- |
- |
PREDICTED: primary amine oxidase [Jatropha curcas] |
| 19 |
Hb_000044_040 |
0.1338421777 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 20 |
Hb_004635_050 |
0.1352102635 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |