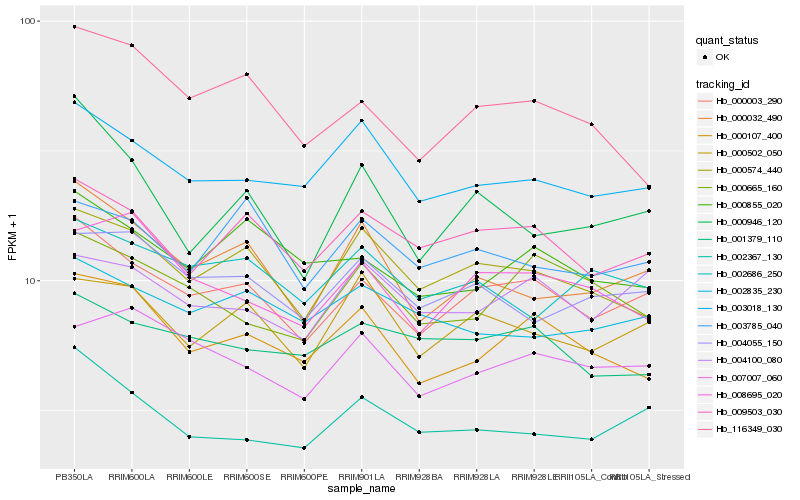
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000003_290 |
0.0 |
- |
- |
hypothetical protein JCGZ_24486 [Jatropha curcas] |
| 2 |
Hb_009503_030 |
0.0711212008 |
- |
- |
protein dimerization, putative [Ricinus communis] |
| 3 |
Hb_000032_490 |
0.077513475 |
- |
- |
PREDICTED: uncharacterized protein At4g10930-like [Jatropha curcas] |
| 4 |
Hb_003018_130 |
0.0801697897 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |
| 5 |
Hb_000574_440 |
0.0802852783 |
- |
- |
PREDICTED: uncharacterized protein LOC105634833 [Jatropha curcas] |
| 6 |
Hb_002367_130 |
0.0843461616 |
- |
- |
UNC-50 family protein isoform 1 [Theobroma cacao] |
| 7 |
Hb_000107_400 |
0.085519198 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46870-like [Jatropha curcas] |
| 8 |
Hb_000502_050 |
0.0856410355 |
transcription factor |
TF Family: HSF |
Heat shock factor protein HSF8, putative [Ricinus communis] |
| 9 |
Hb_004100_080 |
0.0863040019 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000855_020 |
0.0869311766 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X1 [Populus euphratica] |
| 11 |
Hb_004055_150 |
0.0886068955 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001379_110 |
0.0916275739 |
- |
- |
PREDICTED: sec1 family domain-containing protein MIP3 [Jatropha curcas] |
| 13 |
Hb_007007_060 |
0.0920805354 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_116349_030 |
0.0935097169 |
- |
- |
PREDICTED: zinc finger MYND domain-containing protein 15 isoform X3 [Jatropha curcas] |
| 15 |
Hb_003785_040 |
0.0937538465 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_002686_250 |
0.094145828 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 65 kDa protein isoform X1 [Jatropha curcas] |
| 17 |
Hb_000665_160 |
0.0947924219 |
- |
- |
PREDICTED: cysteine--tRNA ligase, cytoplasmic isoform X1 [Jatropha curcas] |
| 18 |
Hb_000946_120 |
0.0949739974 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_008695_020 |
0.0951419782 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 1 [Jatropha curcas] |
| 20 |
Hb_002835_230 |
0.0951748574 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |