| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
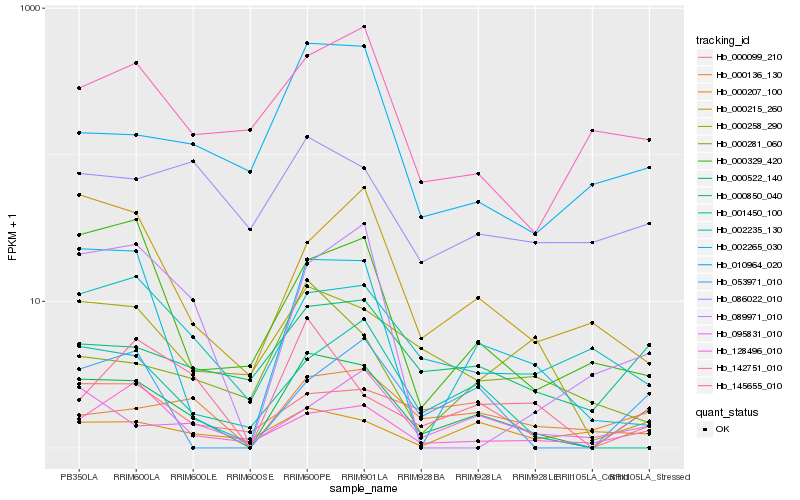
Hb_000522_140 |
0.0 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 2 |
Hb_000850_040 |
0.2379318127 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000099_210 |
0.2446558047 |
- |
- |
PREDICTED: uncharacterized protein LOC105645736 [Jatropha curcas] |
| 4 |
Hb_000136_130 |
0.2468523663 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 7a [Solanum lycopersicum] |
| 5 |
Hb_002235_130 |
0.2620934509 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002265_030 |
0.2629922654 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable calcium-binding protein CML49-like [Glycine max] |
| 7 |
Hb_010964_020 |
0.2635708312 |
- |
- |
RNA helicase [Arabidopsis thaliana] |
| 8 |
Hb_145655_010 |
0.2675524837 |
- |
- |
PREDICTED: lactoylglutathione lyase [Jatropha curcas] |
| 9 |
Hb_001450_100 |
0.2707993859 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 10 |
Hb_095831_010 |
0.2708220834 |
- |
- |
PREDICTED: uncharacterized protein LOC104884649 [Beta vulgaris subsp. vulgaris] |
| 11 |
Hb_000215_260 |
0.2717678083 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: leucine-rich repeat extensin-like protein 4 [Jatropha curcas] |
| 12 |
Hb_128496_010 |
0.2736059782 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_142751_010 |
0.2749781371 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000258_290 |
0.275098909 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP15 [Jatropha curcas] |
| 15 |
Hb_053971_010 |
0.2753241328 |
- |
- |
- |
| 16 |
Hb_000329_420 |
0.2782679509 |
- |
- |
Nuc-1 negative regulatory protein preg, putative [Ricinus communis] |
| 17 |
Hb_000207_100 |
0.2814102393 |
- |
- |
PREDICTED: ABC transporter G family member 25 [Jatropha curcas] |
| 18 |
Hb_086022_010 |
0.2820826576 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1 homolog, mitochondrial [Nicotiana tomentosiformis] |
| 19 |
Hb_000281_060 |
0.2838386013 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_089971_010 |
0.2842157596 |
- |
- |
PREDICTED: (S)-coclaurine N-methyltransferase [Cucumis sativus] |