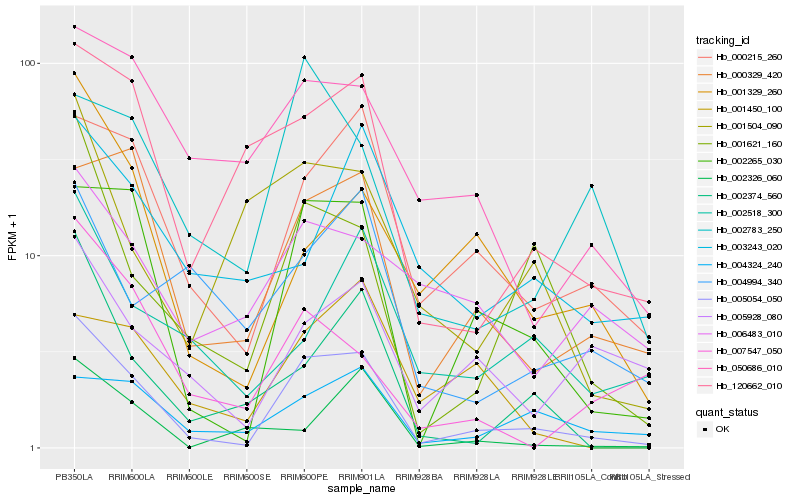
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005054_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 2 |
Hb_002265_030 |
0.1707201326 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable calcium-binding protein CML49-like [Glycine max] |
| 3 |
Hb_000215_260 |
0.1956959326 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: leucine-rich repeat extensin-like protein 4 [Jatropha curcas] |
| 4 |
Hb_001621_160 |
0.219393727 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 5 |
Hb_000329_420 |
0.2246339416 |
- |
- |
Nuc-1 negative regulatory protein preg, putative [Ricinus communis] |
| 6 |
Hb_120662_010 |
0.2283204459 |
- |
- |
hypothetical protein POPTR_0006s18790g [Populus trichocarpa] |
| 7 |
Hb_002374_560 |
0.2426567558 |
- |
- |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH11-like [Populus euphratica] |
| 8 |
Hb_005928_080 |
0.2450813596 |
- |
- |
PREDICTED: probable galacturonosyltransferase 6 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001329_260 |
0.2470555616 |
- |
- |
PREDICTED: uncharacterized protein LOC105642694 isoform X1 [Jatropha curcas] |
| 10 |
Hb_050686_010 |
0.2504391893 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004324_240 |
0.2530169955 |
- |
- |
tetrahydrofolylpolyglutamate synthase, putative [Ricinus communis] |
| 12 |
Hb_002518_300 |
0.2558775799 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001504_090 |
0.2631638116 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_006483_010 |
0.2667433815 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_002783_250 |
0.2703775917 |
- |
- |
gibberellin-regulated protein 1-like precursor [Jatropha curcas] |
| 16 |
Hb_002326_060 |
0.2723636911 |
- |
- |
PREDICTED: pollen-specific leucine-rich repeat extensin-like protein 3 [Populus euphratica] |
| 17 |
Hb_004994_340 |
0.2741696153 |
- |
- |
- |
| 18 |
Hb_007547_050 |
0.274956022 |
- |
- |
Auxin response factor 9 [Aegilops tauschii] |
| 19 |
Hb_001450_100 |
0.2761517258 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 20 |
Hb_003243_020 |
0.2790852153 |
- |
- |
PREDICTED: uncharacterized protein LOC105629849 [Jatropha curcas] |