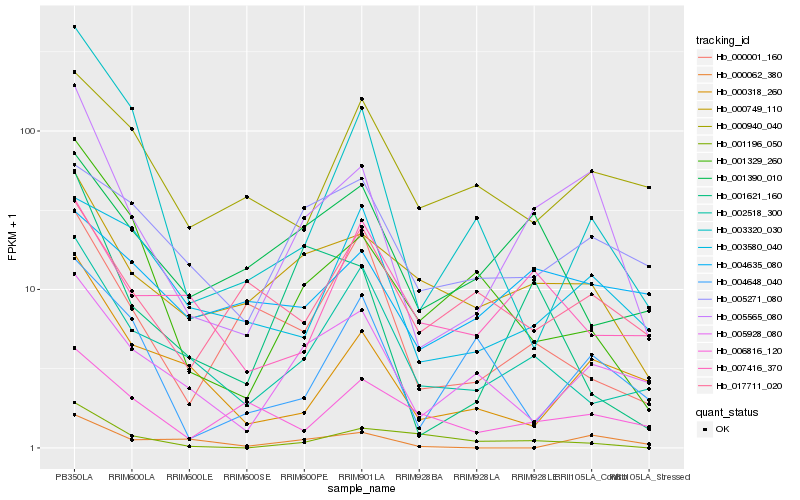
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005565_080 |
0.0 |
- |
- |
PREDICTED: retrotransposon-like protein 1 [Jatropha curcas] |
| 2 |
Hb_000749_110 |
0.2364887549 |
- |
- |
PREDICTED: choline transporter-like protein 2 [Jatropha curcas] |
| 3 |
Hb_000318_260 |
0.2522446852 |
- |
- |
PREDICTED: cell division cycle-associated 7-like protein [Jatropha curcas] |
| 4 |
Hb_005928_080 |
0.25308674 |
- |
- |
PREDICTED: probable galacturonosyltransferase 6 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001621_160 |
0.2572134127 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 6 |
Hb_002518_300 |
0.2634866981 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001329_260 |
0.2734343572 |
- |
- |
PREDICTED: uncharacterized protein LOC105642694 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000062_380 |
0.276001969 |
- |
- |
PREDICTED: putative F-box protein At1g67623 [Jatropha curcas] |
| 9 |
Hb_003580_040 |
0.2842104483 |
- |
- |
PREDICTED: protein ENHANCED DOWNY MILDEW 2-like [Jatropha curcas] |
| 10 |
Hb_003320_030 |
0.2858962319 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001196_050 |
0.2862626176 |
- |
- |
- |
| 12 |
Hb_004648_040 |
0.2869690359 |
- |
- |
PREDICTED: uncharacterized protein LOC105643056 [Jatropha curcas] |
| 13 |
Hb_007416_370 |
0.2872834947 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000001_160 |
0.2874605018 |
- |
- |
PREDICTED: probable glutamate carboxypeptidase 2 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001390_010 |
0.2887126769 |
- |
- |
PREDICTED: uncharacterized protein LOC105635994 [Jatropha curcas] |
| 16 |
Hb_017711_020 |
0.2907108735 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 56 isoform X1 [Jatropha curcas] |
| 17 |
Hb_004635_080 |
0.2907323036 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_005271_080 |
0.2911750198 |
- |
- |
receptor kinase, putative [Ricinus communis] |
| 19 |
Hb_006816_120 |
0.2927537711 |
- |
- |
Nicotianamine synthase, putative [Ricinus communis] |
| 20 |
Hb_000940_040 |
0.2936134713 |
- |
- |
PREDICTED: protein XAP5 CIRCADIAN TIMEKEEPER isoform X1 [Jatropha curcas] |