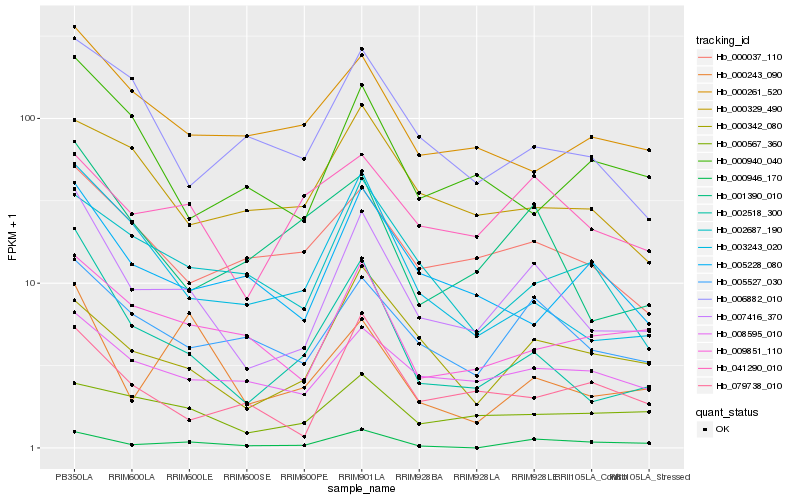
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007416_370 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002518_300 |
0.1384847461 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic [Jatropha curcas] |
| 3 |
Hb_005527_030 |
0.1575189507 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g42630 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003243_020 |
0.1775696372 |
- |
- |
PREDICTED: uncharacterized protein LOC105629849 [Jatropha curcas] |
| 5 |
Hb_008595_010 |
0.1799505105 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g06710, mitochondrial [Jatropha curcas] |
| 6 |
Hb_000342_080 |
0.1857205009 |
- |
- |
hypothetical protein L484_019524 [Morus notabilis] |
| 7 |
Hb_009851_110 |
0.186221 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 8 |
Hb_000037_110 |
0.1876705821 |
- |
- |
PREDICTED: cation/H(+) antiporter 18-like [Populus euphratica] |
| 9 |
Hb_001390_010 |
0.1890710568 |
- |
- |
PREDICTED: uncharacterized protein LOC105635994 [Jatropha curcas] |
| 10 |
Hb_000261_520 |
0.1976720071 |
- |
- |
PREDICTED: uncharacterized protein LOC105633192 [Jatropha curcas] |
| 11 |
Hb_000946_170 |
0.1976782266 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase-like [Jatropha curcas] |
| 12 |
Hb_041290_010 |
0.198490596 |
- |
- |
thioredoxin family protein [Populus trichocarpa] |
| 13 |
Hb_006882_010 |
0.1993311218 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Cucumis sativus] |
| 14 |
Hb_005228_080 |
0.2005210556 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 3-I [Jatropha curcas] |
| 15 |
Hb_079738_010 |
0.2012715605 |
- |
- |
hypothetical protein CISIN_1g036582mg, partial [Citrus sinensis] |
| 16 |
Hb_002687_190 |
0.2015142879 |
- |
- |
prolyl oligopeptidase [Cicer arietinum] |
| 17 |
Hb_000567_360 |
0.2030298997 |
- |
- |
hypothetical protein CISIN_1g004656mg [Citrus sinensis] |
| 18 |
Hb_000940_040 |
0.2031210667 |
- |
- |
PREDICTED: protein XAP5 CIRCADIAN TIMEKEEPER isoform X1 [Jatropha curcas] |
| 19 |
Hb_000329_490 |
0.2044754737 |
- |
- |
hypothetical protein JCGZ_14582 [Jatropha curcas] |
| 20 |
Hb_000243_090 |
0.2060485976 |
- |
- |
PREDICTED: protein RTE1-HOMOLOG isoform X1 [Jatropha curcas] |