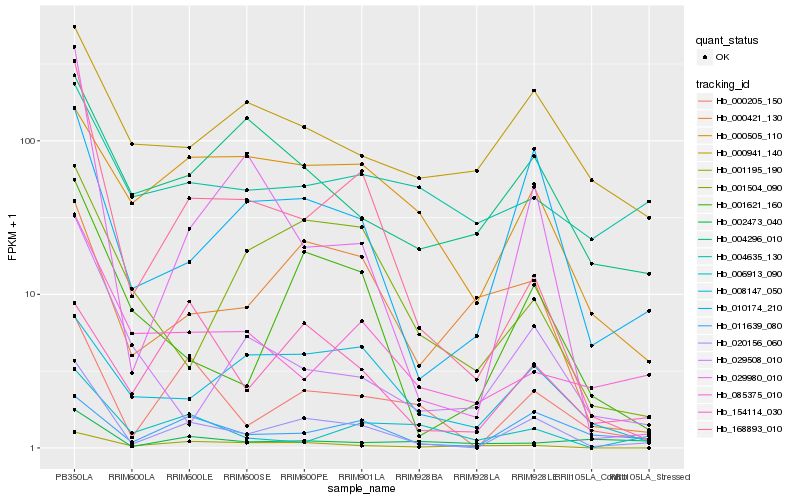
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020156_060 |
0.0 |
- |
- |
PREDICTED: ankyrin-1-like [Prunus mume] |
| 2 |
Hb_000205_150 |
0.1982991051 |
- |
- |
PREDICTED: uncharacterized protein LOC105647752 [Jatropha curcas] |
| 3 |
Hb_010174_210 |
0.2081582118 |
- |
- |
PREDICTED: small heat shock protein, chloroplastic-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_168893_010 |
0.2197291789 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Nelumbo nucifera] |
| 5 |
Hb_001621_160 |
0.233875025 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 6 |
Hb_029980_010 |
0.2600758723 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Jatropha curcas] |
| 7 |
Hb_011639_080 |
0.2633969766 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: receptor-like protein kinase FERONIA [Populus euphratica] |
| 8 |
Hb_085375_010 |
0.2697663403 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_000421_130 |
0.2757408794 |
transcription factor |
TF Family: C2C2-Dof |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_001195_190 |
0.2776914151 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000941_140 |
0.2816957591 |
- |
- |
PREDICTED: activator of 90 kDa heat shock protein ATPase homolog 1 [Jatropha curcas] |
| 12 |
Hb_029508_010 |
0.2830494867 |
transcription factor |
TF Family: HSF |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_004296_010 |
0.2914265576 |
- |
- |
PREDICTED: activator of 90 kDa heat shock protein ATPase homolog 1 [Jatropha curcas] |
| 14 |
Hb_006913_090 |
0.2915060205 |
- |
- |
PREDICTED: uncharacterized protein LOC101215996 [Cucumis sativus] |
| 15 |
Hb_001504_090 |
0.2939661959 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_154114_030 |
0.2942196107 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 17 |
Hb_000505_110 |
0.2951871692 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump [Jatropha curcas] |
| 18 |
Hb_008147_050 |
0.2989309507 |
- |
- |
PREDICTED: uncharacterized protein LOC105646845 [Jatropha curcas] |
| 19 |
Hb_002473_040 |
0.3051894408 |
- |
- |
PREDICTED: rRNA methyltransferase 3B, mitochondrial [Jatropha curcas] |
| 20 |
Hb_004635_130 |
0.3068759042 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein G [Populus euphratica] |