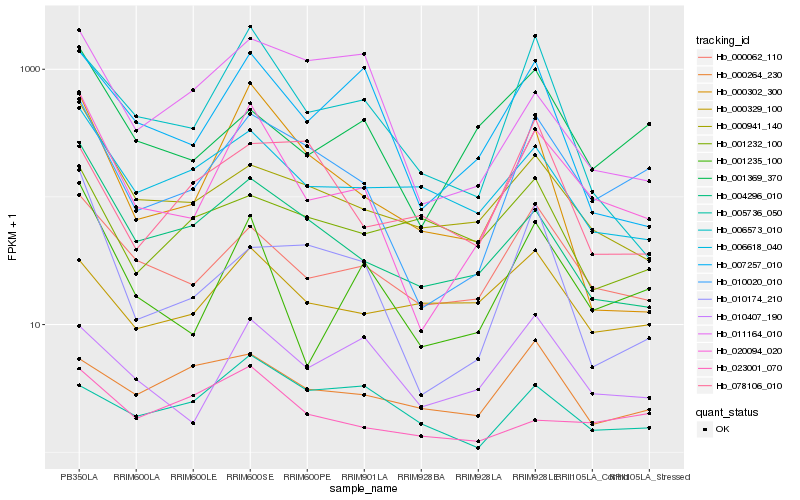
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010020_010 |
0.0 |
- |
- |
cytosolic class II low molecular weight heat shock protein [Prunus dulcis] |
| 2 |
Hb_020094_020 |
0.1579277823 |
- |
- |
PREDICTED: 17.1 kDa class II heat shock protein-like [Pyrus x bretschneideri] |
| 3 |
Hb_000062_110 |
0.1857301402 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_17894 [Jatropha curcas] |
| 4 |
Hb_004296_010 |
0.214287269 |
- |
- |
PREDICTED: activator of 90 kDa heat shock protein ATPase homolog 1 [Jatropha curcas] |
| 5 |
Hb_001369_370 |
0.2170718125 |
- |
- |
PREDICTED: 23.6 kDa heat shock protein, mitochondrial [Jatropha curcas] |
| 6 |
Hb_010174_210 |
0.2178853536 |
- |
- |
PREDICTED: small heat shock protein, chloroplastic-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_010407_190 |
0.2214315334 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 8 |
Hb_000941_140 |
0.2222530353 |
- |
- |
PREDICTED: activator of 90 kDa heat shock protein ATPase homolog 1 [Jatropha curcas] |
| 9 |
Hb_078106_010 |
0.2236584021 |
- |
- |
seed maturation protein PM37 [Populus trichocarpa] |
| 10 |
Hb_000264_230 |
0.2244877808 |
- |
- |
PREDICTED: uncharacterized protein LOC105649528 [Jatropha curcas] |
| 11 |
Hb_006618_040 |
0.2258519014 |
- |
- |
PREDICTED: hsp70-Hsp90 organizing protein 3-like [Jatropha curcas] |
| 12 |
Hb_001235_100 |
0.2278638746 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 13 |
Hb_006573_010 |
0.2339927771 |
- |
- |
heat shock protein [Hevea brasiliensis] |
| 14 |
Hb_000329_100 |
0.2340951007 |
- |
- |
PREDICTED: uncharacterized protein LOC105643144 [Jatropha curcas] |
| 15 |
Hb_005736_050 |
0.2350607014 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like [Jatropha curcas] |
| 16 |
Hb_023001_070 |
0.2350873654 |
- |
- |
PREDICTED: dnaJ protein ERDJ3A-like [Eucalyptus grandis] |
| 17 |
Hb_011164_010 |
0.2399386758 |
- |
- |
cytosolic class II low molecular weight heat shock protein [Prunus dulcis] |
| 18 |
Hb_000302_300 |
0.2412239124 |
- |
- |
PREDICTED: heat shock protein 83 [Populus euphratica] |
| 19 |
Hb_001232_100 |
0.2438601674 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 20 |
Hb_007257_010 |
0.2445614813 |
- |
- |
cytosolic class II low molecular weight heat shock protein [Prunus dulcis] |