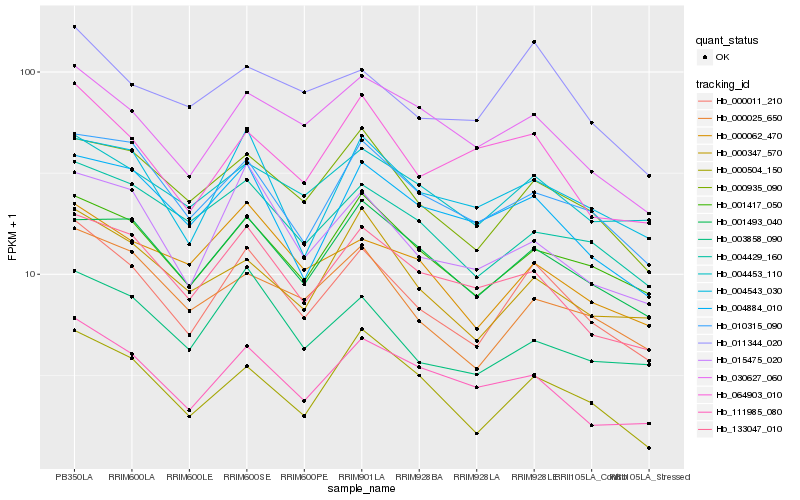
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000011_210 |
0.0 |
- |
- |
PREDICTED: FACT complex subunit SPT16-like [Jatropha curcas] |
| 2 |
Hb_001417_050 |
0.0885834446 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |
| 3 |
Hb_000935_090 |
0.0906721151 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 4 |
Hb_015475_020 |
0.0946911209 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 5 |
Hb_133047_010 |
0.095567923 |
- |
- |
xanthine dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_010315_090 |
0.0983215595 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 7 |
Hb_000025_650 |
0.0983454611 |
- |
- |
exosome complex exonuclease rrp45, putative [Ricinus communis] |
| 8 |
Hb_111985_080 |
0.0990714402 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g19720 [Jatropha curcas] |
| 9 |
Hb_030627_060 |
0.1010021914 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 10 |
Hb_000504_150 |
0.1022609968 |
- |
- |
PREDICTED: nuclear pore complex protein GP210 [Jatropha curcas] |
| 11 |
Hb_001493_040 |
0.1024640664 |
- |
- |
PREDICTED: FACT complex subunit SPT16-like [Jatropha curcas] |
| 12 |
Hb_003858_090 |
0.1029020171 |
- |
- |
Filamin-A-interacting protein, putative [Ricinus communis] |
| 13 |
Hb_004429_160 |
0.1033264238 |
- |
- |
PREDICTED: serine/threonine-protein kinase TAO3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000347_570 |
0.1043809294 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12b isoform X1 [Jatropha curcas] |
| 15 |
Hb_011344_020 |
0.1046608838 |
- |
- |
PREDICTED: hsp70-Hsp90 organizing protein 3-like [Jatropha curcas] |
| 16 |
Hb_004543_030 |
0.1057659086 |
- |
- |
PREDICTED: FIP1[V]-like protein [Jatropha curcas] |
| 17 |
Hb_000062_470 |
0.1061484809 |
- |
- |
PREDICTED: cyclin-dependent kinase G-2 [Jatropha curcas] |
| 18 |
Hb_004453_110 |
0.1062373378 |
- |
- |
Peptidyl-prolyl cis-trans isomerase CYP19-2 isoform 1 [Theobroma cacao] |
| 19 |
Hb_064903_010 |
0.1068083509 |
- |
- |
PREDICTED: T-complex protein 1 subunit delta [Jatropha curcas] |
| 20 |
Hb_004884_010 |
0.1073201023 |
- |
- |
PREDICTED: pre-mRNA-splicing factor CWC22 homolog [Jatropha curcas] |