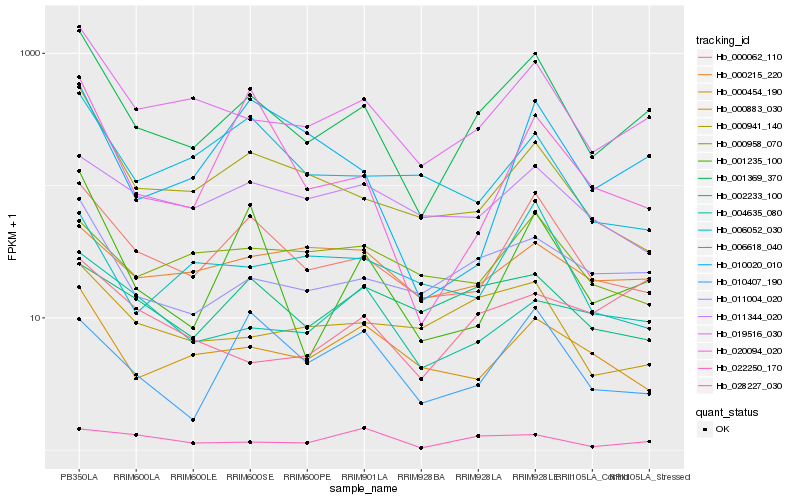
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001369_370 |
0.0 |
- |
- |
PREDICTED: 23.6 kDa heat shock protein, mitochondrial [Jatropha curcas] |
| 2 |
Hb_019516_030 |
0.1280343327 |
- |
- |
Calcyclin-binding protein, putative [Ricinus communis] |
| 3 |
Hb_000062_110 |
0.1434847633 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_17894 [Jatropha curcas] |
| 4 |
Hb_011004_020 |
0.1539219989 |
- |
- |
PREDICTED: pyridoxal 5'-phosphate synthase-like subunit PDX1.2 [Jatropha curcas] |
| 5 |
Hb_000941_140 |
0.1876319297 |
- |
- |
PREDICTED: activator of 90 kDa heat shock protein ATPase homolog 1 [Jatropha curcas] |
| 6 |
Hb_001235_100 |
0.1918274143 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 7 |
Hb_000454_190 |
0.2007728622 |
- |
- |
PREDICTED: probable ribosome-binding factor A, chloroplastic [Jatropha curcas] |
| 8 |
Hb_004635_080 |
0.2028794512 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_000883_030 |
0.2029636247 |
- |
- |
PREDICTED: uncharacterized protein LOC105636683 [Jatropha curcas] |
| 10 |
Hb_010020_010 |
0.2170718125 |
- |
- |
cytosolic class II low molecular weight heat shock protein [Prunus dulcis] |
| 11 |
Hb_010407_190 |
0.2201104294 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 12 |
Hb_002233_100 |
0.2204110272 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 6-like [Malus domestica] |
| 13 |
Hb_028227_030 |
0.2205416784 |
- |
- |
PREDICTED: uncharacterized protein LOC105644817 isoform X1 [Jatropha curcas] |
| 14 |
Hb_006052_030 |
0.2225001132 |
- |
- |
PREDICTED: dnaJ protein ERDJ3A [Jatropha curcas] |
| 15 |
Hb_000215_220 |
0.2245892746 |
- |
- |
PREDICTED: derlin-1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_011344_020 |
0.224736996 |
- |
- |
PREDICTED: hsp70-Hsp90 organizing protein 3-like [Jatropha curcas] |
| 17 |
Hb_020094_020 |
0.2258498433 |
- |
- |
PREDICTED: 17.1 kDa class II heat shock protein-like [Pyrus x bretschneideri] |
| 18 |
Hb_022250_170 |
0.2268995529 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g26782, mitochondrial-like [Populus euphratica] |
| 19 |
Hb_006618_040 |
0.2271073801 |
- |
- |
PREDICTED: hsp70-Hsp90 organizing protein 3-like [Jatropha curcas] |
| 20 |
Hb_000958_070 |
0.2286719607 |
- |
- |
hypothetical protein PRUPE_ppa007681mg [Prunus persica] |