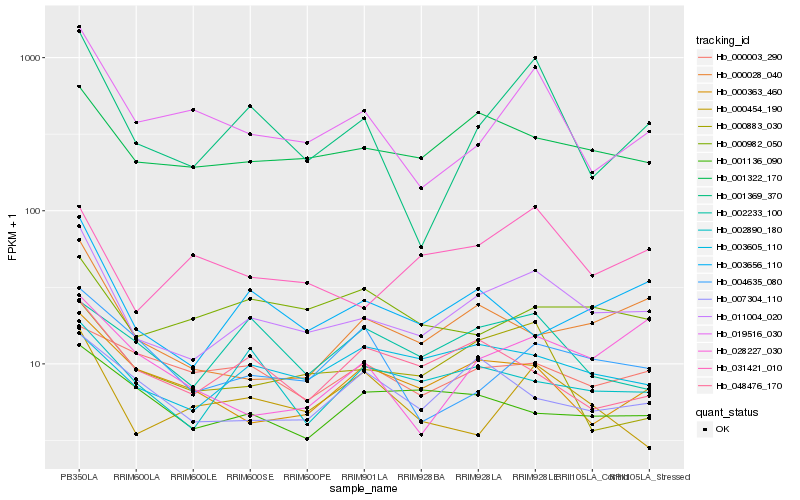
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011004_020 |
0.0 |
- |
- |
PREDICTED: pyridoxal 5'-phosphate synthase-like subunit PDX1.2 [Jatropha curcas] |
| 2 |
Hb_001322_170 |
0.1287498606 |
- |
- |
PREDICTED: bax inhibitor 1-like [Jatropha curcas] |
| 3 |
Hb_000028_040 |
0.1519274281 |
- |
- |
PREDICTED: prostaglandin E synthase 2-like [Jatropha curcas] |
| 4 |
Hb_001369_370 |
0.1539219989 |
- |
- |
PREDICTED: 23.6 kDa heat shock protein, mitochondrial [Jatropha curcas] |
| 5 |
Hb_003656_110 |
0.1547271139 |
- |
- |
phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 6 |
Hb_000883_030 |
0.159558885 |
- |
- |
PREDICTED: uncharacterized protein LOC105636683 [Jatropha curcas] |
| 7 |
Hb_019516_030 |
0.1609963998 |
- |
- |
Calcyclin-binding protein, putative [Ricinus communis] |
| 8 |
Hb_031421_010 |
0.1628213003 |
- |
- |
PREDICTED: copper chaperone for superoxide dismutase, chloroplastic/cytosolic [Jatropha curcas] |
| 9 |
Hb_000363_460 |
0.1630052946 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial isoform X1 [Jatropha curcas] |
| 10 |
Hb_002890_180 |
0.1660004971 |
- |
- |
PREDICTED: squamous cell carcinoma antigen recognized by T-cells 3 [Jatropha curcas] |
| 11 |
Hb_048476_170 |
0.1705894949 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 3-II-like [Citrus sinensis] |
| 12 |
Hb_000454_190 |
0.1712627959 |
- |
- |
PREDICTED: probable ribosome-binding factor A, chloroplastic [Jatropha curcas] |
| 13 |
Hb_007304_110 |
0.173564053 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase 1 isoform X3 [Jatropha curcas] |
| 14 |
Hb_000982_050 |
0.1757817539 |
- |
- |
PREDICTED: uncharacterized protein LOC105643124 isoform X1 [Jatropha curcas] |
| 15 |
Hb_003605_110 |
0.1786827718 |
- |
- |
PREDICTED: putative 1-phosphatidylinositol-3-phosphate 5-kinase FAB1D isoform X1 [Jatropha curcas] |
| 16 |
Hb_004635_080 |
0.1788712244 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 17 |
Hb_001136_090 |
0.1793184463 |
- |
- |
Protein PNS1, putative [Ricinus communis] |
| 18 |
Hb_028227_030 |
0.1793409953 |
- |
- |
PREDICTED: uncharacterized protein LOC105644817 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000003_290 |
0.1809333547 |
- |
- |
hypothetical protein JCGZ_24486 [Jatropha curcas] |
| 20 |
Hb_002233_100 |
0.1810585633 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 6-like [Malus domestica] |